Method for enriching DNA target regions by synergistic PCR method based on artificial nucleic acid analogue technology
An enrichment and nucleotide technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve problems such as uneven amplification products, difficulty in primer design, and difficulty in optimizing amplification conditions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
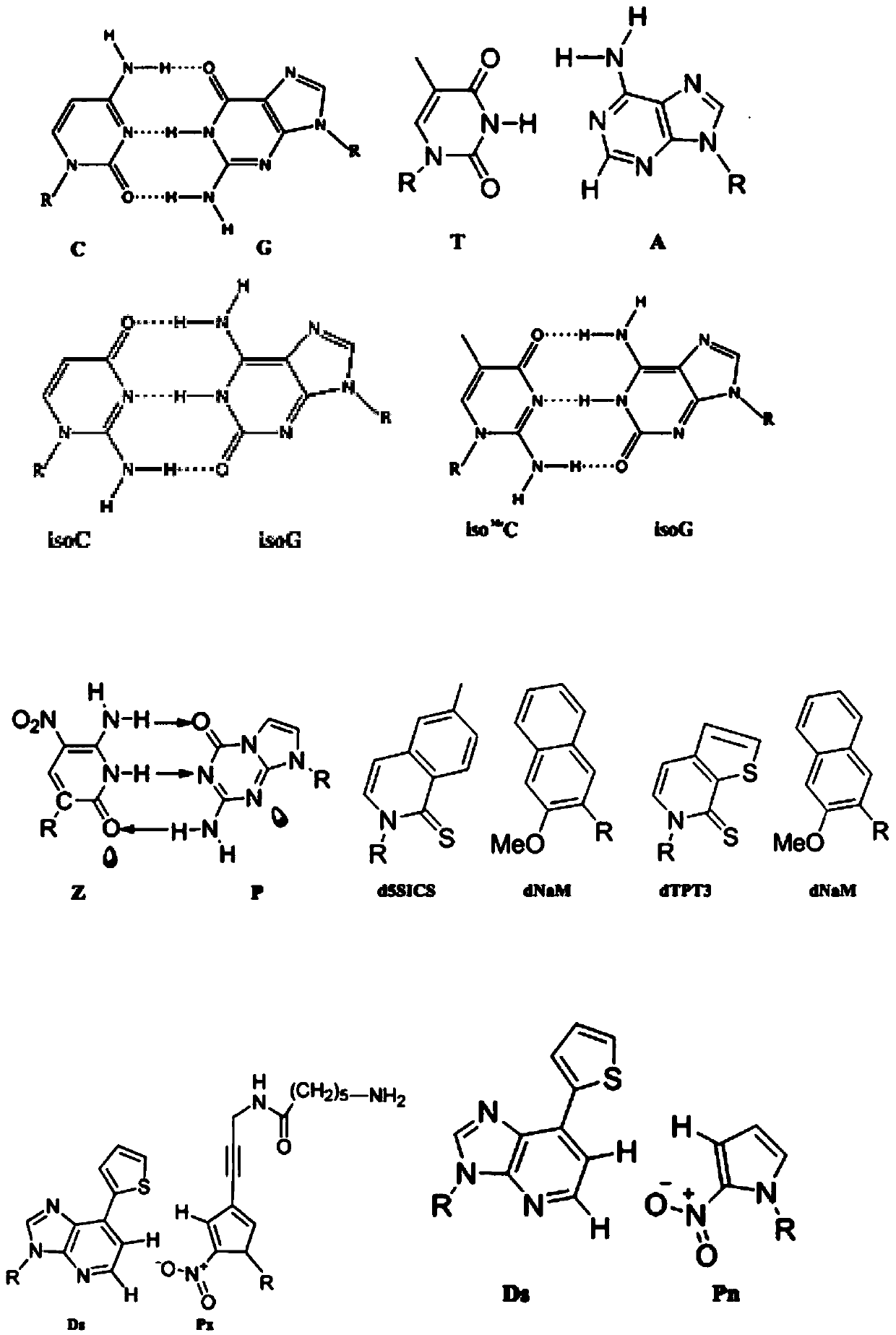
[0079] The design of embodiment 1, ANABB-PCR primer
[0080] In this embodiment, the partial sequence of the human genome TP53 gene (this partial sequence is shown in sequence 1 of the sequence listing, 2530bp) is selected as an example, the primer is designed, and the specific effect of the primer is verified in Example 2 and Example 3 / implementation In Example 4, the effect of primers on the amplification of complex templates was verified.
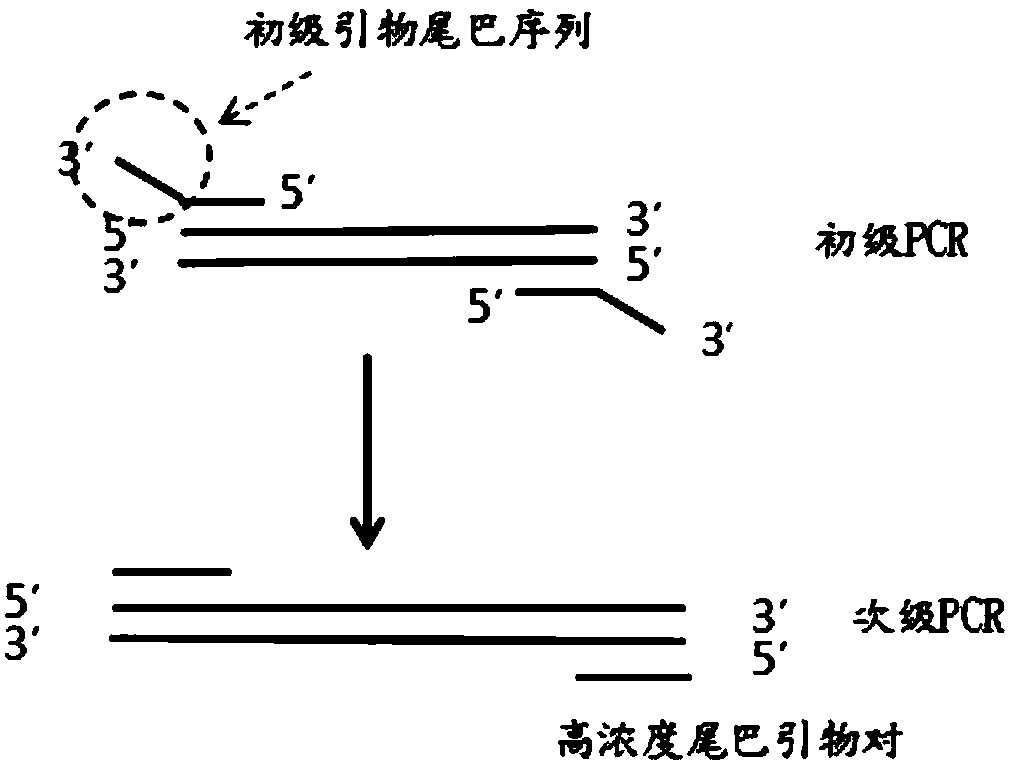
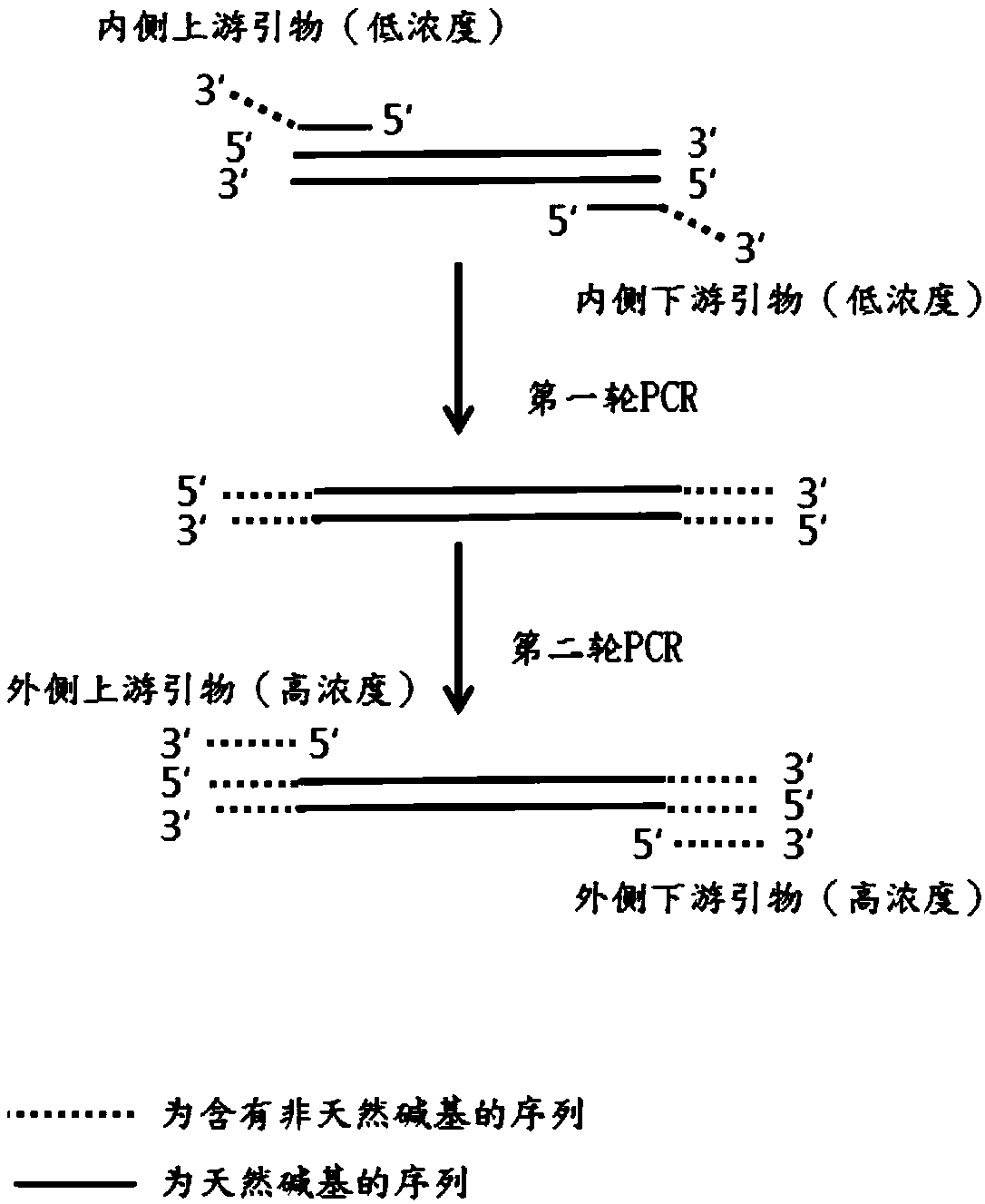
[0081] The natural base primer set consists of a natural base inner primer pair and a natural base outer primer pair. The primer pair inside the natural base is composed of primer F1 (upstream primer inside the natural base) and primer R1 (downstream primer inside the natural base). The primer pair outside the natural base is composed of primer F2 (upstream primer outside the natural base) and primer R2 (downstream primer outside the natural base).
[0082] The ANABB primer set consists of ANABB inner primer pair and ANABB outer primer...
Embodiment 2
[0083] Embodiment 2, the specificity of non-natural base primer
[0084] Primers to be tested: primer 1, primer 2 (introduced 2 Ds), primer 3 (introduced 3 Ds), primer 4 (introduced 4 Ds).
[0085] 1. Comparison of primer self-dimer effects
[0086] Carry out the following test for each primer to be tested respectively.
[0087] Reaction system (pH7.0, 20μl): from 10μl 480SYBR Green I Master, 1 μl primer solution to be tested and ddH 2 O composition. In the reaction system, the concentration of the primer to be tested is 0.2 μM.
[0088] After the reaction system was prepared, real-time fluorescence quantitative PCR was performed.
[0089] Reaction program: pre-denaturation at 95°C for 10 min; 40 cycles of denaturation at 95°C for 10 s, annealing at 55°C for 20 s, and extension at 72°C for 60 s; fluorescence signals were collected at 72°C.
[0090] see results Figure 4 . Introducing non-natural nucleotides into primers can effectively avoid the formation of primer se...
Embodiment 3
[0098] Embodiment 3, the amplification effect of ANABB primer set to complex template
[0099] In this example, human genomic DNA was used as a template, and each primer set in Example 1 was used to perform PCR amplification on a 2530 bp longer template of the TP53 gene, so as to verify the effect of the ANABB primer set on complex template amplification.
[0100] 1. Amplification of ANABB primer set (test set)
[0101] The first round of PCR reaction: reaction system (pH7.0, 20 μl): by 10 μl 480SYBR GreenIMaster, 0.1 μl primer F3 solution, 0.1 μl primer R3 solution, 1 μl template solution, 0.5 μl dPxTP solution and ddH 2 O composition. In 1 μl template solution, the genomic DNA content is 100ng. In the reaction system, the concentration of primer F3 was 0.02 μM, the concentration of primer R3 was 0.02 μM, and the concentration of dPxTP was 0.5 mM. Reaction program: pre-denaturation at 95°C for 10 minutes; denaturation at 95°C for 10 s, annealing at 60°C for 20 s, extensi...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap