Primer composition and reagent for fusion gene detection
A technology of primer composition and fusion gene, which is applied in the field of primer composition and fusion gene detection reagents, can solve the problems of inability to directly detect fusion gene fragments, long operation time, low sensitivity, etc., and achieve good application prospects and low overall cost , the effect of high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] 1. Sample preparation
[0033] The total RNA of each cell line shown in the table below was extracted and quantified by Qubit.
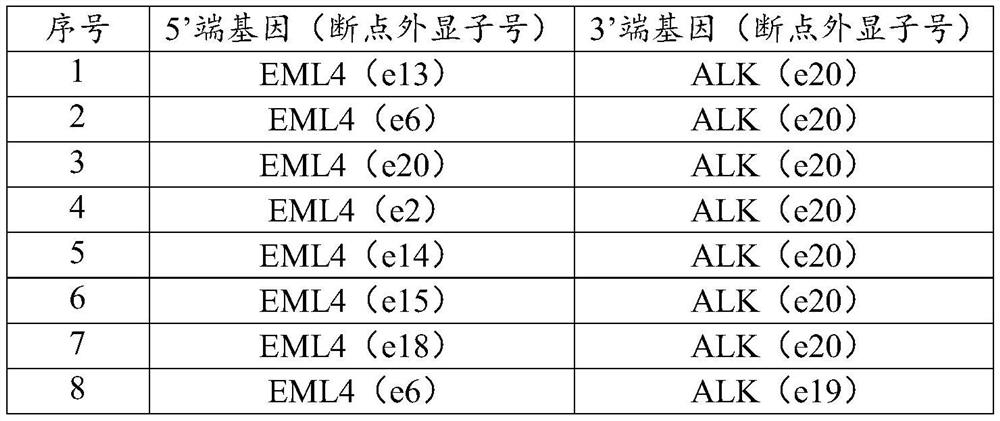
[0034] cell line fusion form H2228 EML4-ALK KM12 TPM3-NTRK1 LC-2 / ad CCDC6-RET HCC78 SLC34A2-ROS1 A549 none (female)
[0035] 2. Experimental process:
[0036] (1) Prepare the qPCR reaction solution as shown in the table below, and add 4 kinds of RNA samples to each tube.
[0037]
[0038] (2) Cover the tube cap, mix by inverting to avoid air bubbles, and then centrifuge quickly.
[0039] (3) Run the qPCR reaction according to the procedure in the table.
[0040]
[0041]
[0042] 3. Test results
[0043] (1) The following table shows the Ct value detected by tube 1, and U.D in the table: Undetermined.
[0044] Fluorescence channel FAM HEX Texas Red Cy5 Corresponding gene ALK NTRK1 NRG housekeeping gene H2228-1 27.79 U.D. U.D. 25.66 H2228...
Embodiment 2
[0052] 1. Sample preparation
[0053] The total RNA of cell lines H2228 and HCC78 were extracted and quantified by Qubit.
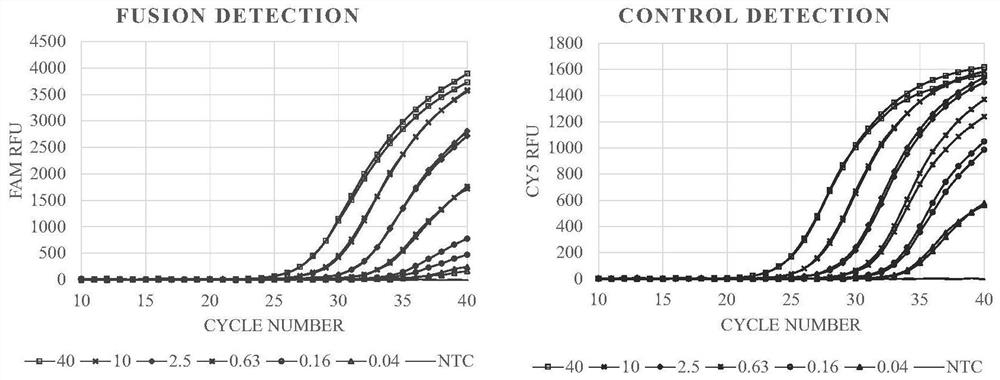
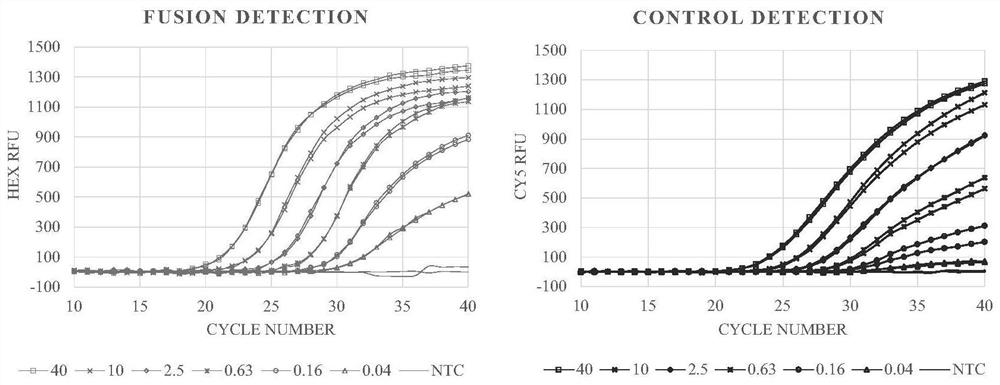
[0054] The total RNA of the two cell lines was serially diluted 4 times to form a series of solutions containing 40, 10, 2.5, 0.63, 0.16, 0.04 ng RNA per 4 μL respectively.
[0055]2. Experimental process
[0056] (1) Prepare the qPCR reaction solution as shown in the table below, and add 2 series of RNA solutions as templates to each tube (only the corresponding fusion form is detected for each RNA).
[0057]
[0058]
[0059] (2) Cover the tube cap, mix by inverting to avoid air bubbles, and then centrifuge quickly.
[0060] (3) Run the qPCR reaction according to the procedure in the table.
[0061]
[0062] 3. Test results
[0063] (1) Data list: See the table below for the detected Ct value.
[0064] Fluorescence channel FAM HEX Cy5 Corresponding gene ALK ROS1 housekeeping gene H2228-40ng 25.54 / 23.01 ...
Embodiment 3
[0070] 1. Sample preparation
[0071] RNA was extracted from 5 clinical FFPE samples and quantified by Qubit.
[0072] 2. Experimental process
[0073] (1) Prepare the qPCR reaction solution as shown in the table below, and add 50 ng of each clinical sample to each tube.
[0074]
[0075] (2) Cover the tube cap, mix by inverting to avoid air bubbles, and then centrifuge quickly.
[0076] (3) Run the qPCR reaction according to the procedure in the table.
[0077]
[0078] 3. Test results
[0079] (1) See the table below for the Ct value detected by tube 1.
[0080] Fluorescence channel FAM HEX Texas Red Cy5 Corresponding gene ALK NTRK1 NRG housekeeping gene SMH 28.34 U.D. U.D. 26.14 WW U.D. U.D. 25.47 19.23 YZG U.D. U.D. U.D. 29.17 WBJ U.D. U.D. 27.17 20.53 YYH U.D. U.D. U.D. 29.69
[0081] (2) See the table below for the Ct value detected by tube 2.
[0082] Fluore...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com