Double centromere distorted chromosome analysis and prediction method based on multi-scale fusion method
A multi-scale fusion and prediction method technology, applied in image analysis, neural learning methods, image enhancement, etc., can solve problems such as poor robustness and poor detection accuracy, achieve fast analysis speed, small error, and improve analysis efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
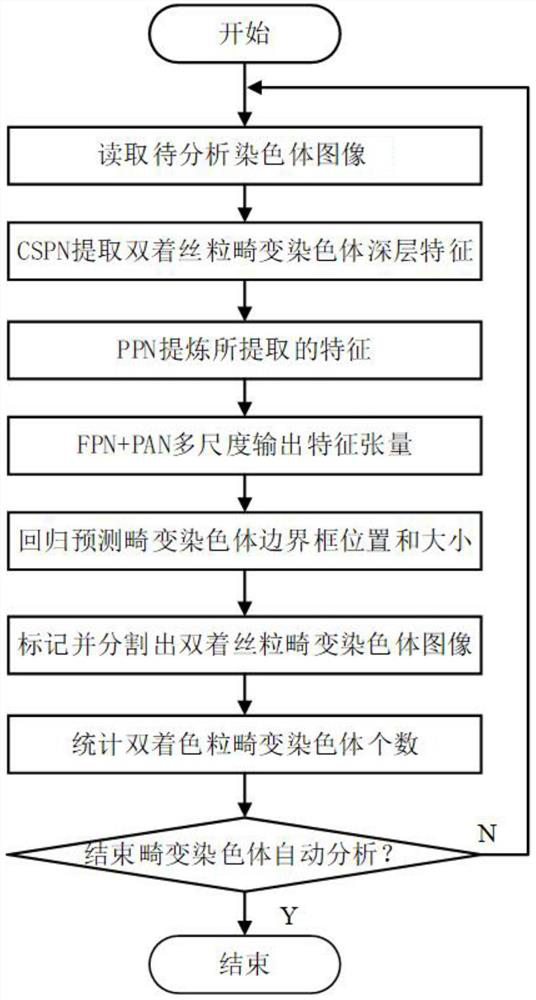
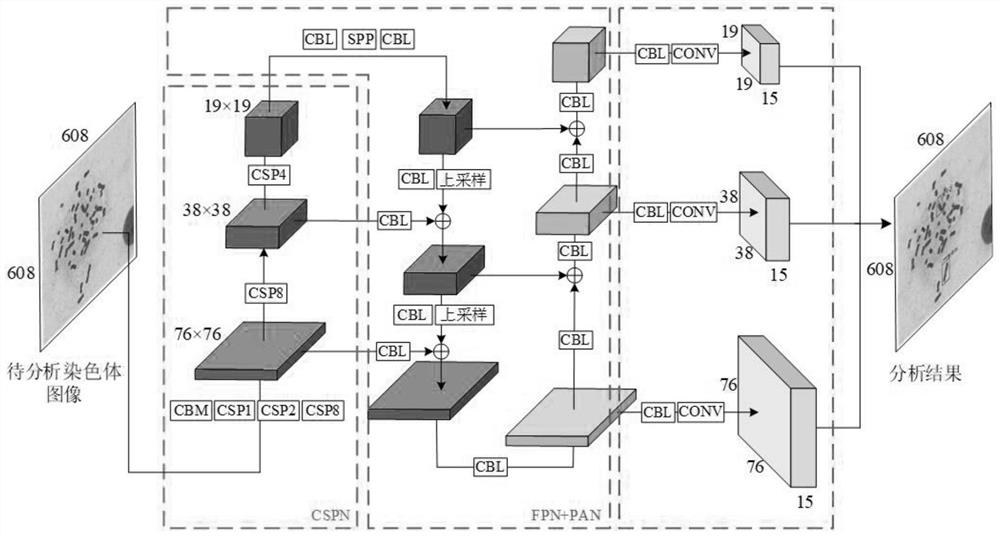
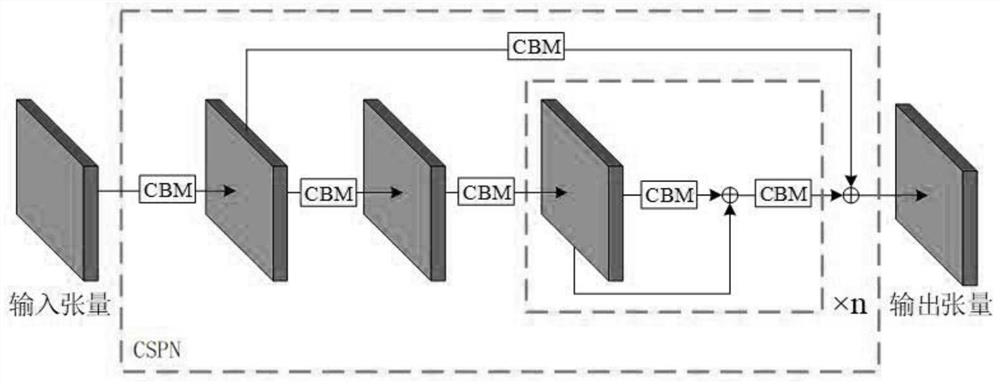
[0044] like Figure 1-2 As shown, the network structure of the designed neural network model is mainly composed of three parts: the backbone neural network CSPN, the FPN+PAN feature fusion network and the output prediction end. The backbone neural network CSPN is mainly composed of several CSPs, and the specific backbone neural network CSPN is composed of five cross-stage partial neural network modules (Cross Stage Partial, CSP). CSP module structure such as image 3 As shown, the CSP module is composed of several CBM modules and n-cycle residual units. The CBM module mainly performs convolution operations on the input feature map and then uses the Mish activation function to process the features.
[0045] The invention provides a method for analyzing and predicting dicentric aberration chromosomes based on a multi-scale fusion method. Before performing regression prediction on the dicentric aberration chromosomes, a neural network model is trained. The training steps are as ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com