Methods for detecting and treating cancers having adenosine pathway activation
A cancer, genetic technology for the detection and treatment of cancers with adenosine pathway activation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
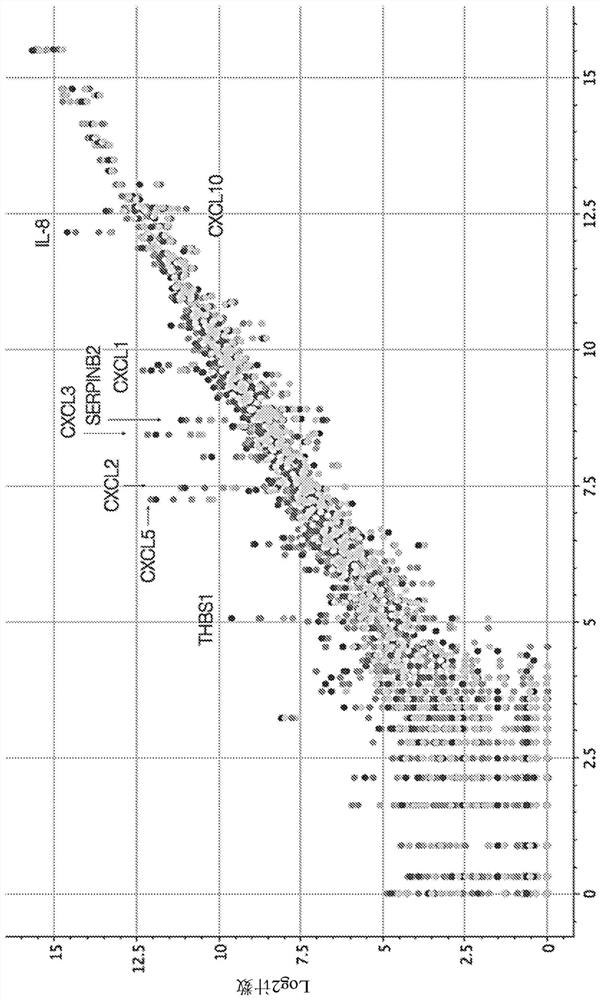
[0217] Example 1: mRNA expression in PBMC is regulated by NECA
[0218] Peripheral cells from normal healthy humans treated with 5'-N-ethylcarboxamidoadenosine (NECA) and activated with anti-CD3 / CD28 antibodies were analyzed by NanoString hybridization-based quantification (NanoString Technologies, Inc.). Purified RNA from blood mononuclear cells (PBMC). figure 1 NanoString quantitative data are shown; marked genes were consistently regulated by NECA treatment. Each point represents the expression level of a specific gene. Dot shading indicates the concentration of NECA and / or different PBMC donors used. Genes above the diagonal axis are upregulated relative to DMSO control. Genes below the diagonal axis are downregulated relative to DMSO control. Table 3 provides a list of genes up- or down-regulated by 0.1 μM, 1 μM and / or 10 μM NECA treatment (compared to vehicle-treated controls) as determined by NanoString NPR1 sequencing.
[0219] Table 3. NanoString NECA Response ...
example 2
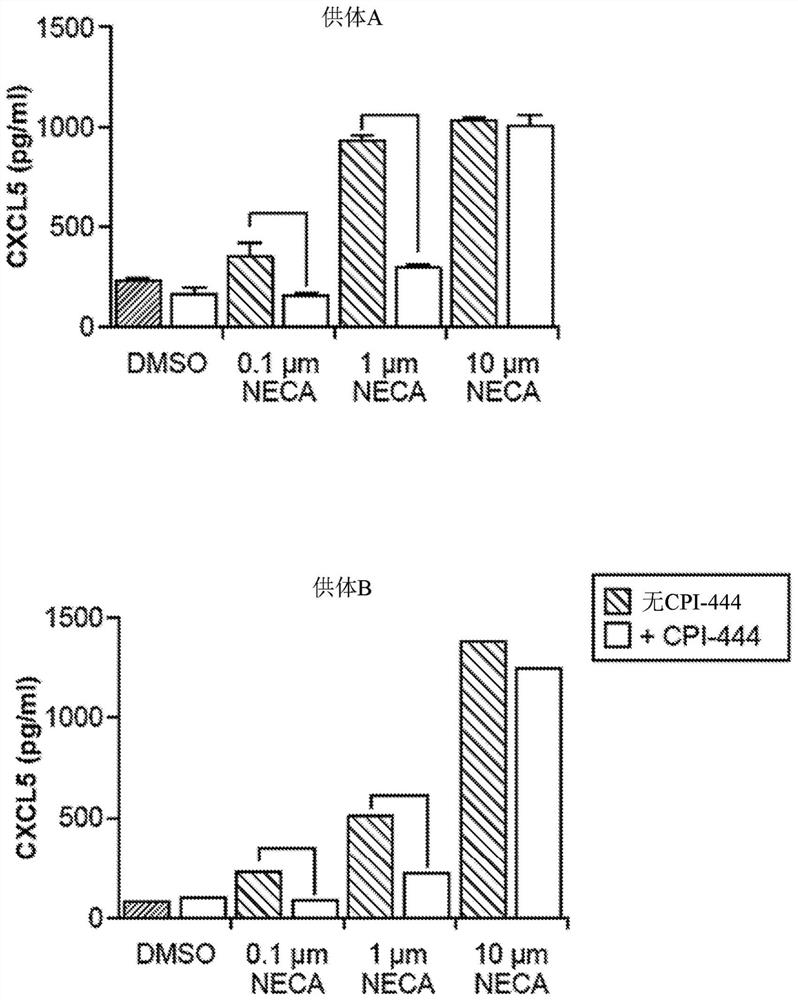
[0231] Example 2: CPI-444 inhibits NECA-induced cytokine expression
[0232] Activation of CXCL5 protein expression by NECA was assessed. PBMCs were collected from two different human donors. like Figure 2AAs indicated, cells were treated with NECA followed by CPI-444 and then anti-CD3 / CD28 to activate PBMCs. CPI-444 (Corvus Pharmaceuticals) is an antagonist of the adenosine A2A receptor. Two days after activation, culture supernatants were assessed for CXCL5 protein by ELISA. exist Figure 2B and 2C provided data. Activation of CXCL5 expression by NECA. This activation in CXCL5 could be blocked by the addition of CPI-444, an A2AR receptor antagonist that neutralizes the immunosuppressive effect of NECA. This demonstrates that induction of CXCL5 is specific for signaling through adenosine receptors.
example 3
[0233] Example 3: CCL20 expression correlates with expression of a subset of adenosine-regulated genes in multiple tumor types
[0234] Using the Cancer Genome Atlas (National Cancer Institute, National Human Genome Research Institute); available at Genomic Data Commons Data Portal cancergenome. nih.gov)), the gene expression of CCL20 compared to the expression of a subset of genes exhibiting regulation by adenosine was determined in various solid tumor types. For each tumor type, the expression level of the adenosine pathway was calculated as the mean of Log2 showing the expression of the genes induced by adenosine (CXCL1, CXCL2, CXCL3, CXCL5, SERPINB2, IL8 and IL1B). Gene expression of CCL20 is highly correlated with expression of this subset of genes in solid tumors ( Figure 3A and 3B ), thus suggesting that CCL20 (and co-regulated genes) can be used as a surrogate for the expression of this subset of genes. p<0.0001 for all tumor types. This group of genes represented...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com