Metalloenzyme active site comparison method based on pharmacophore and alpha-carbon characteristics
An active site, metalloenzyme technology, applied in genomics, used to analyze two-dimensional or three-dimensional molecular structure, instruments, etc., can solve the problem that metalloenzymes are difficult to achieve the desired effect, and the metal ion characteristics of metalloenzymes are rarely considered. And other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
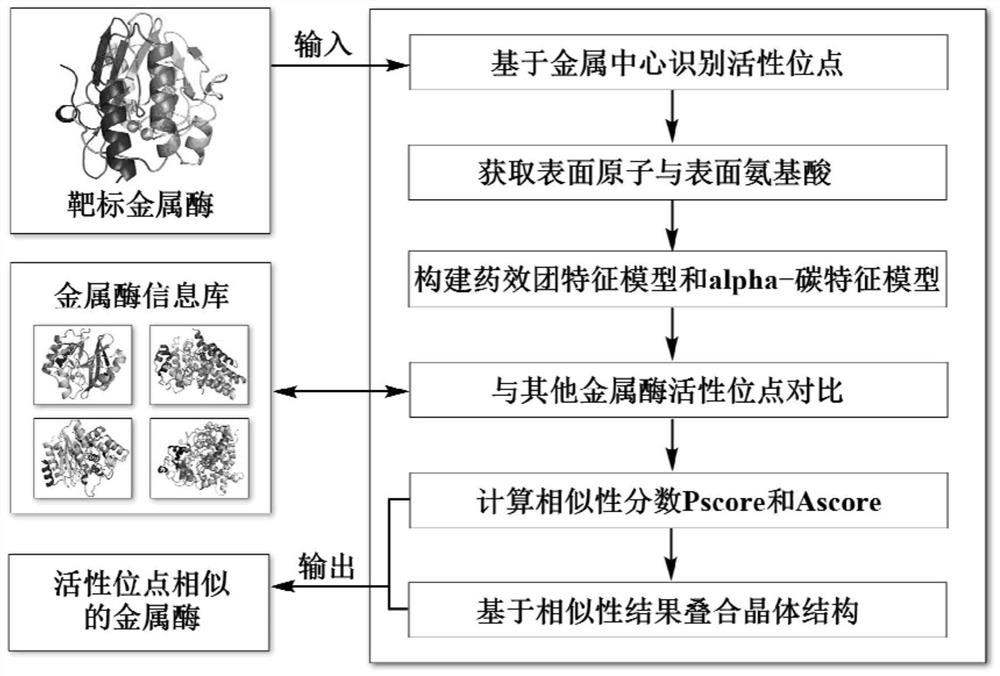
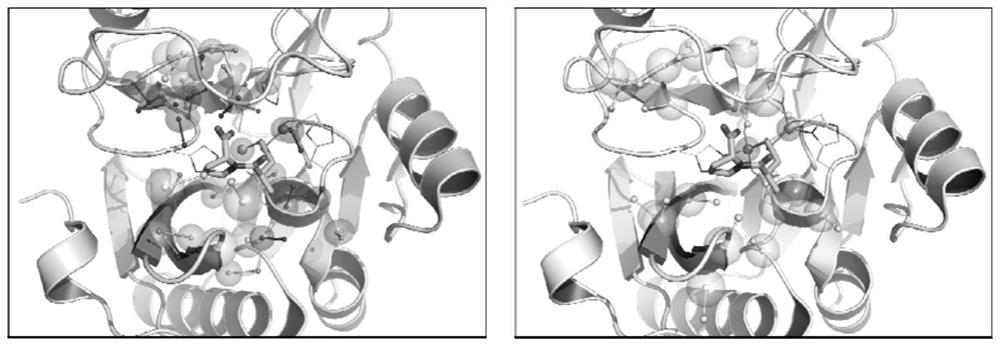
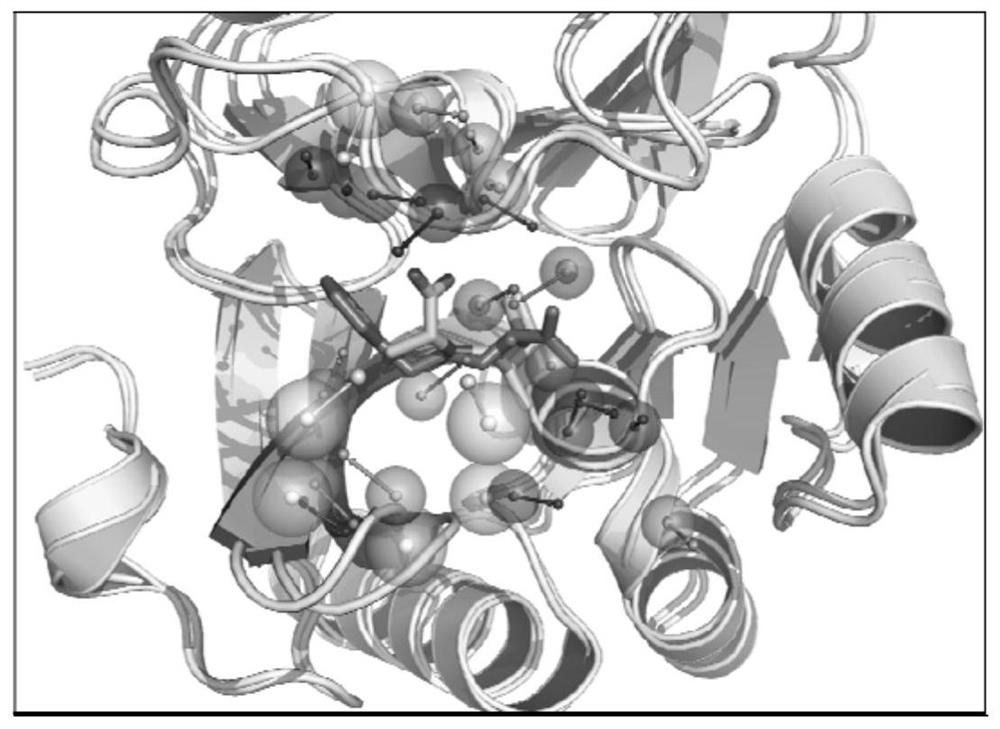
[0046] attached figure 1 The workflow of a metalloenzyme active site alignment method based on pharmacophore and alpha-carbon features is described. Input the crystal structure of the target metalloenzyme, identify its active site based on the principle of metal center combined with solvent accessibility, obtain the surface atoms and surface amino acids of the site, and construct the corresponding pharmacophore characteristic model and alpha- carbon characteristic model; then compare the similarity with the characteristic models of other metalloenzyme active sites in the metal information library, and calculate the similarity score Pscore based on the pharmacophore characteristic and the similarity score based on the alpha-carbon characteristic according to the matching results of the characteristic model. The similarity score Ascore, and superimpose the crystal structure of the target metalloenzyme and the similar metalloenzyme according to the similarity results; combine Psc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com