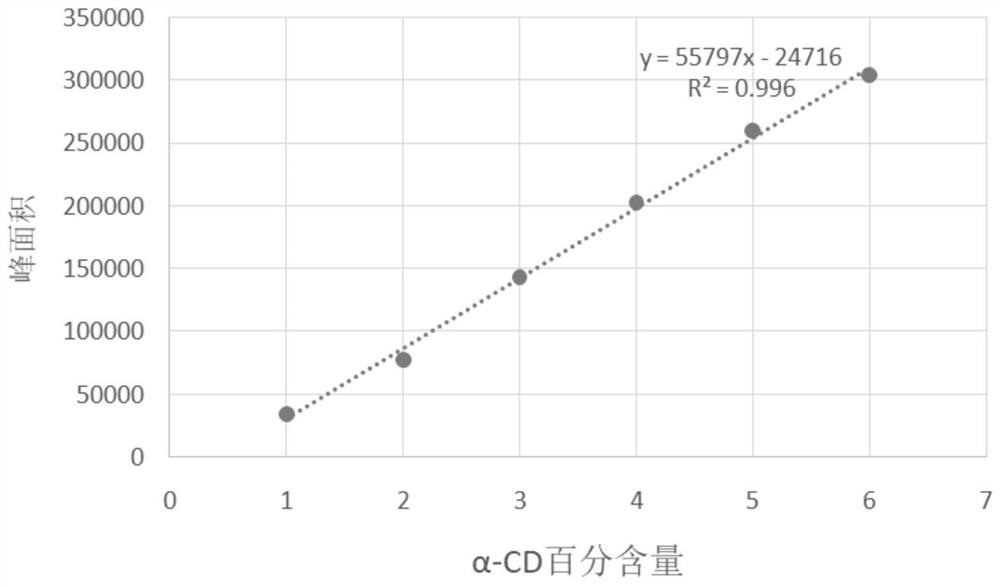
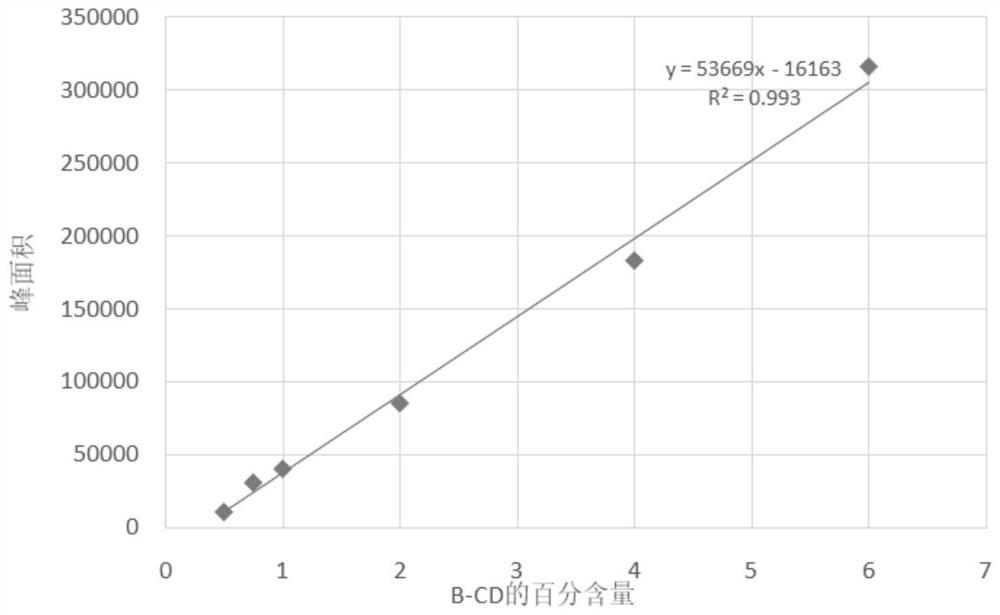
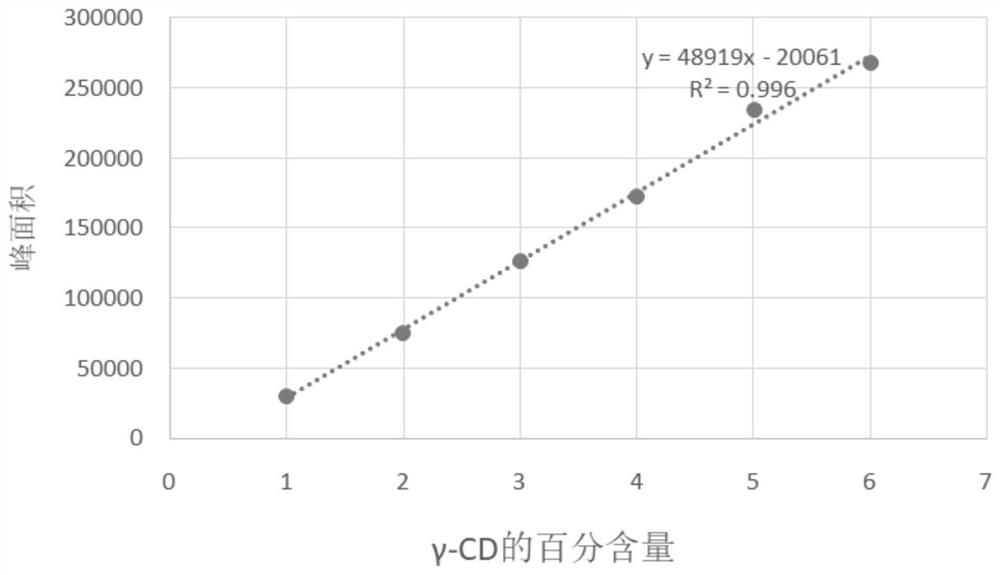
Application of protein CGTase as cyclodextrin glycosyltransferase
A glycosyltransferase, protein technology, applied in the direction of glycosyltransferase, transferase, application, etc., can solve the problems of small production scale, limited production and application of γ-CD, and high price
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0096] Embodiment 1, the discovery of protein CGTase1 and its 15 mutant proteins
[0097] 1. Discovery of the CGTase1 gene
[0098] The CGTase1 gene shown in SEQ ID NO: 2 was artificially synthesized.
[0099] The CGTase1 gene encodes the protein CGTase1 shown in SEQ ID NO:1.
[0100] 2. Obtaining Mutant Proteins
[0101] The inventors of the present invention carried out saturation mutation at position 248 of protein CGTase1, and obtained 15 mutant proteins including deletion mutations. details as follows:
[0102] (1) Deleting the 742-744 nucleotides (GCA) from the 5' end of SEQ ID NO: 2 to obtain double-stranded DNA molecule 1. The protein encoded by double-stranded DNA molecule 1 is named protein 1.
[0103] That is, protein 1 is obtained after the alanine at position 248 of protein CGTase1 is deleted.
[0104] (2) Replace the 742nd-744th nucleotides (GCA) of SEQ ID NO: 2 from the 5' end with CGA to obtain double-stranded DNA molecule 2. The protein encoded by the d...
Embodiment 2
[0132] Example 2. Application of the protein CGTase1 and its muteins as cyclodextrin glycosyltransferases in the production of α-CD, β-CD and γ-CD
[0133] 1. Construction of recombinant plasmid 1-recombinant plasmid 16
[0134] 1. Artificially synthesize the double-stranded DNA molecule 1-double-stranded DNA molecule 15 in Step 2 of Example 1 and the double-stranded DNA molecule shown in SEQ ID NO: 2 (named double-stranded DNA molecule 16).
[0135] 2. Using double-stranded DNA molecule 1 as a template, use primer S2: 5'-CGC GGATCC ATGATTCGAAGGCTTTC-3' (the underline is the recognition sequence of restriction endonuclease BamHI) and primer A3: 5'-CGG CTCGAG A primer pair consisting of TTGATTGTAATTCACTTC-3' (the underline is the recognition sequence of the restriction endonuclease XhoI) was used for PCR amplification to obtain a PCR amplification product of about 2000 bp.
[0136] 3. Digest the PCR amplified product obtained in step 2 with restriction endonucleases BamHI a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com