Method for evaluating, detecting and/or predicting beef quality traits of consumption cattle
A technology for quality traits and beef, applied in biochemical equipment and methods, microbiological determination/inspection, DNA/RNA fragments, etc., can solve problems such as unreported research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1 4
[0057] Example 1 Amplification of Sichuan Yak ACOT12 Gene SNP Site
[0058] 1. Experimental samples
Embodiment 1
[0059] The 102 Sichuan yaks in Example 1 came from a cattle farm in Aba Prefecture, Sichuan Province. The blood samples of healthy yaks in the same environment, the same feeding conditions, and the same period were collected and stored at -20°C for the extraction of blood genome DNA and the collection of related meat Quality trait data and analysis.
[0060] 2. Experimental method
[0061] 1. DNA extraction
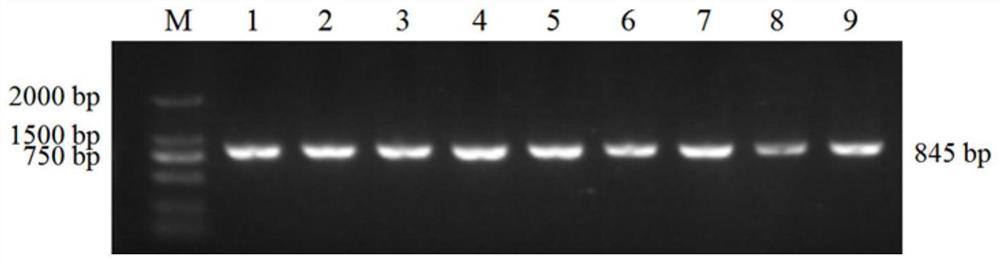
[0062] According to the instructions, the blood genome DNA extraction kit was used to extract DNA from Sichuan yak blood samples, the concentration and purity of DNA were detected by micro-spectrophotometer, and the quality of DNA was detected by agarose gel electrophoresis.
[0063] 2. Primer design
[0064] According to the bovine ACOT12 gene sequence published in Ensembl (ENSBTAT00000014753.6), a pair of specific amplification primers was designed and synthesized by Sangon Bioengineering (Shanghai) Co., Ltd.
[0065] The sequences of the specific amplification prime...
Embodiment 2
[0080] Example 2 Population genetic characteristics of 3 SNPs loci of ACOT12 gene
[0081] 1. Experimental method
[0082] All 102 Sichuan yak sequencing results obtained in Example 1 were analyzed, using Popgene 32 software to calculate the gene frequency, genotype frequency, genetic heterozygosity (H), effective allele number (Ne) of alleles; PIC0.6 software calculates polymorphic information content (PIC).
[0083] 2. Experimental results
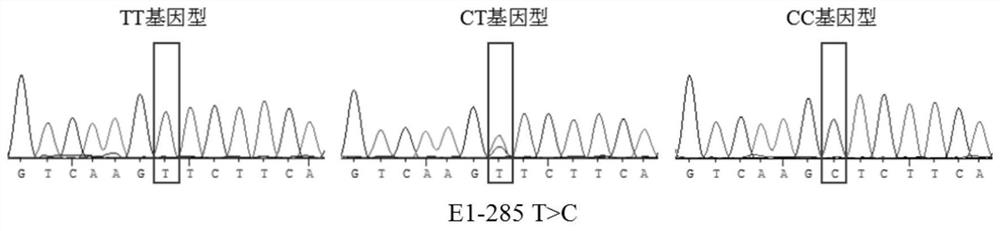
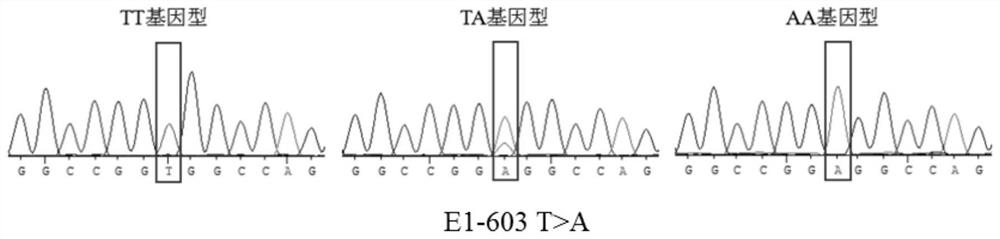
[0084] The population genetic characteristics of the three SNPs of the ACOT12 gene are shown in Table 1. It can be seen from Table 1 that the dominant genotype in SNP site 1 (E1-285T>C), SNP site 2 (E1-603 T>A), and SNP site 3 (E1-675 C>G) is TC , TA, CC, the dominant alleles are T, A, C, respectively, and the frequency is greater than 0.5.
[0085] The genetic heterozygosity (H) and polymorphic information content (PIC) of SNP locus 1 (E1-285 T>C) and SNP locus 2 (E1-603 T>A) were between 0.25 and 0.50, It shows that the above loci...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com