Assembly method and assembly device for chromosome level genome
An assembly method and chromosome technology, applied in the field of genome assembly, can solve problems such as difficult to achieve genome assembly at the chromosome level
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] In this embodiment, a genome assembly method at the chromosome level is provided, such as figure 1 As shown, the assembly method includes:
[0030] S101, obtaining the known chromosome-level genome of the same species as a reference genome;
[0031] S103, comparing the contig or scaffold of the individual to be assembled with the reference genome to obtain corresponding coordinate information;
[0032] S105. Mount the contig or scaffold of the individual to be assembled to the chromosome level according to the coordinate information, and obtain the genome of the individual to be assembled at the chromosome level.
[0033] This assembly method uses the published chromosome-level genome of the same species as the reference genome, and compares the contig or scaffold-level genome of the newly tested individual (for example, using mummer) to the reference genome, so as to realize the reference of the newly tested individual. The purpose of mounting to the chromosome level...
Embodiment 2
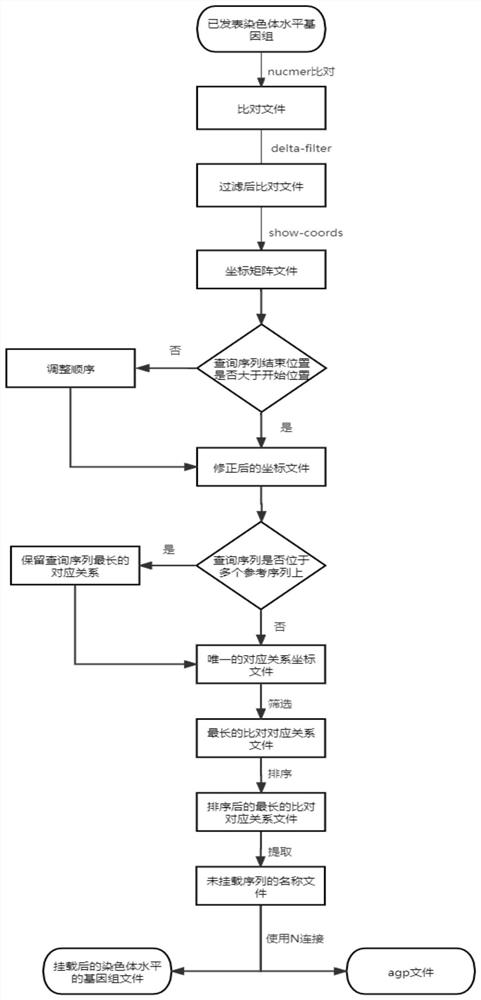
[0048] This embodiment provides a method for mounting genome parameters to the chromosome level, such as figure 2 As shown, the specific implementation process is as follows:
[0049] 1. Use the nucmer command in mummer to compare the target genome to the chromosome-level genome, the reference sequence is the chromosome-level genome sequence, and the target genome sequence is the query sequence to obtain the comparison file.
[0050]2. Use the delta-filter command in mummer to filter the shorter alignment sequences in the alignment file to obtain the filtered alignment file.
[0051] 3. Use the show-coords command in mummer to convert the filtered alignment file into an aligned coordinate matrix file.
[0052] 4. According to the coordinate matrix file after mummer comparison, output the corrected coordinate file. Determine whether the end position of the query sequence in the comparison region is greater than the start position of the query sequence in the comparison regio...
Embodiment 3
[0064] This embodiment provides a chromosome-level genome assembly device, the assembly device includes: an acquisition module, a comparison module and a mounting module, wherein,
[0065] The obtaining module is configured to obtain the known chromosome-level genome of the same species as a reference genome;
[0066] The comparison module is configured to compare the contigs or scaffolds of the individuals to be assembled with the reference genome to obtain corresponding coordinate information;
[0067] The mounting module is configured to mount the contig or scaffold of the individual to be assembled to the chromosome level according to the coordinate information, and obtain the genome of the individual to be assembled at the chromosome level.
[0068] Optionally, the alignment module includes a mummer module.
[0069] In a preferred embodiment, the comparison module includes: a mummer module, a correction module and a deduplication module, wherein,
[0070] The mummer mod...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com