Culture medium for directionally inducing and differentiating pluripotent stem cells into hepatocytes as well as culture method and application
A technology of pluripotent stem cells and induced differentiation, applied in the field of directional induction and differentiation of pluripotent stem cells into hepatocytes, which can solve the problems of unsatisfactory cell integration efficiency and low maturity of iHeps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0090] Example 1 sgRNA sequence and donor plasmid construction
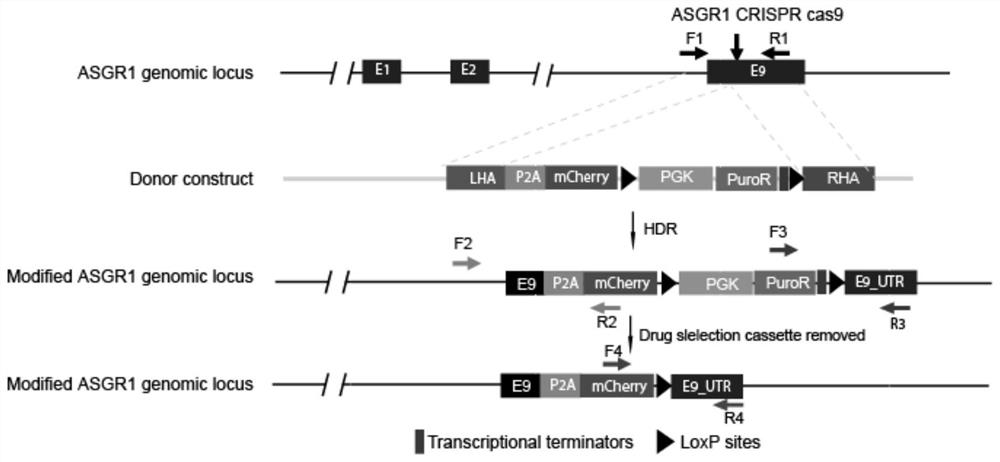
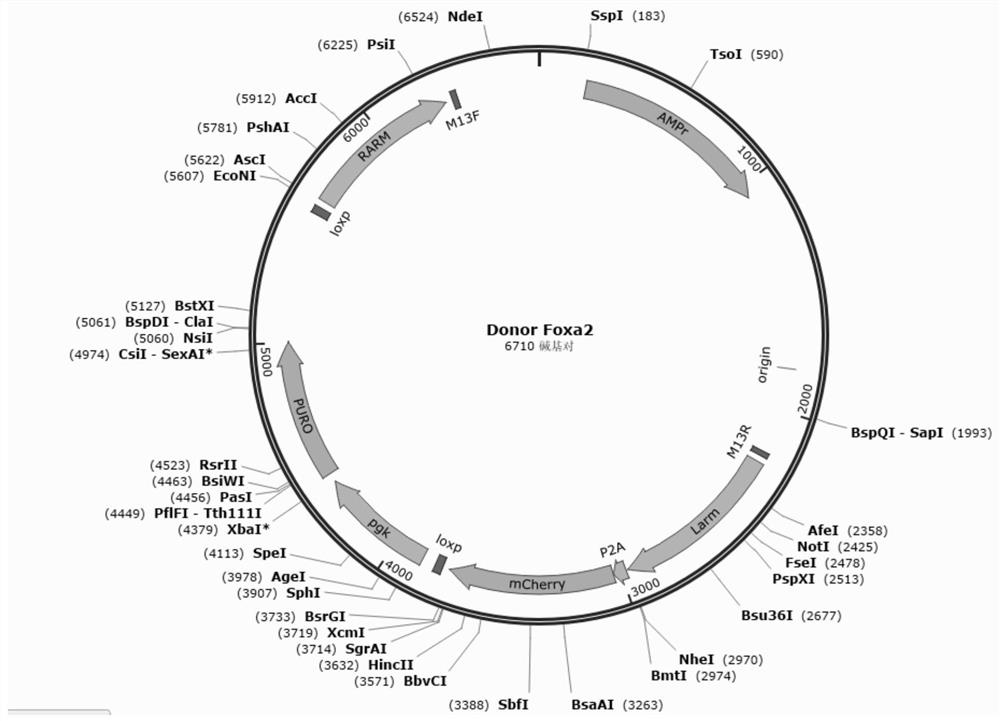
[0091] Donor construct (Donor construct) design see figure 2 . Donor plasmids include LHA-P2A-mCherry-loxP-PGK-PuroR-loxP-RHA; where LHA is the left homology arm, P2A is a self-cleaving 2A peptide, mCherry is a fluorescent protein, and loxP is the loxP site Point, PGK is the mammalian promoter derived from the phosphoglycerate kinase gene, PuroR is the puromycin resistance gene pac, and RHA is the right homology arm.
[0092] according to figure 2 Design vector, insert P2A-mCherry-loxP-PGK-PuroR-loxP expression frame before ASGR1 stop codon, design process is as follows: design left and right arm (LHA, RHA) primer ASGR1-LARM-PF: CTGgaattcCTCCACGTGAAGCAGTTCGT, as SEQ ID NO: 1; ASGR1-LARM-PR: GCGgctagcAAGGAGAGGTGGCTCCTGGCTG, as shown in SEQ ID NO: 2, ASGR1-RARM-PF: GTGgctagcGTCggcgcgccTTTATTTCTTCAATGCCTCGACCTGC, as shown in SEQ ID NO: 3; ASGR1-RARM-PR: GTGaagcttCACTCAACAANOATTCGGGTGGT, as shown in SEQ ID : Sh...
Embodiment 2
[0103] Example 2 ASGR1-mCherry cell line construction
[0104] 1) The first step of screening
[0105] 2.5ug ASGR1 targeting vector and 2.5ug ASGR1 CRISPR-Cas9 vector were transfected into 800,000 hiPSCs by nucleofection, and plated into one well of a 6-well plate. On Day 4, 0.05-1 mg / ml puromycin was added until 99% of the cells died. On Day 5, the puromycin was removed and the mTeSR1 medium was replaced, and the clones were identified when they grew to a suitable size, and the primers F1 / R1 were identified: AGACGGGCTTCAAGTGAGTG (as shown in SEQ ID NO:13) / CCTGCTTCCGAATCCCCAAT (as shown in SEQ ID NO:14), F2 / R2:GGATGTAGGGCTGACCTCGTT (as shown in SEQ ID NO:15) / ACAGCTTCAAGTAGTCGGGG (as shown in SEQ ID NO:16) , F3 / R3: GGTGCATGACCCGCAAGCCC (shown in SEQ ID NO: 17) / TTTGTTGCTTCTGTTCCGCAG (shown in SEQ ID NO: 18). Such as Figure 5 , the target clone was screened by Junction PCR, and the No. 20 single-cell-derived clone with both alleles (alleles) modified was selected for the next...
Embodiment 3
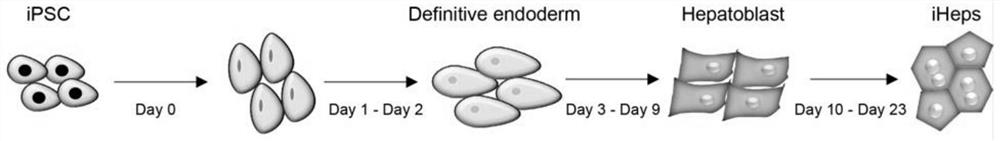
[0108] Example 3 Hepatocyte (iHeps) Differentiation
[0109] In this example, the hiPSCs (20#-13#) obtained in Example 2 were differentiated into iHeps through three stages. 1) In the first stage, hiPSCs were treated with RPMI1640+Activin A (100ng / mL) and WNT3α (20ng / mL) for 24 hours (Day 0), and then the first stage medium without WNT3α was used for the next 2 days (Day1-Day2). In this way, Definitiveendoderm (DE) can be induced for three days. 2) The second stage is to induce DE into hepatoblasts. This stage uses the second stage medium to culture from day3 to day9. The second stage medium composition: 80% knockout DMEM (KO-DMEM), 20% knockout serum replacement (KSR) , 0.5% GlutaMAX, 1% non-essential amino acids (NEAA), 0.1 mM beta-mercaptoethanol and 1% DMSO. 3) Subsequently, on Day 10, the medium S3 of the third stage was replaced and cultured for 7 days or more. S3 medium contained hepatoZYME-SFM, 1% GlutaMAX, 10 μM hydrocortisone 21-hemisuccinatesodium salt (HCC), 20....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com