Adenylate cyclase containing FYVE structural domain as well as coding gene and application thereof
A technology of adenylate cyclase and coding gene, which is applied in the field of pathogenic oomycete novel adenylate cyclase and its coding gene and application, and can solve the problems of soybean seed rot, economic loss, rot, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Example 1, Cloning of the adenylyl cyclase PsZFAC gene containing the FYVE domain in Phytophthora sojae
[0057] 1.1 Phytophthora soybean total RNA extraction
[0058] Phytophthora sojae strain P6497 was cultivated in the dark at 25°C for 4 days on 10% V8 solid medium, and the bacteria cake with a diameter of 5 mm was picked from the edge of the colony and inoculated in 10% V8 liquid medium, and harvested after 4 days of dark culture at 25°C For mycelium, take about 30mg of mycelium, put it into a 2mL mold-free and sterile centrifuge tube, add two steel balls with a diameter of 5mm, freeze it in liquid nitrogen, and use a ball mill to grind it to powder. The SV Total RNA Extraction Kit (Z3100) from Promega Company was used to extract Phytophthora sojae RNA, and the specific method steps refer to the description of plant tissue RNA extraction in the kit.
[0059] 1.2 Synthesis of first-strand cDNA by reverse transcription of Phytophthora sojae
[0060] First-strand cDN...
Embodiment 2
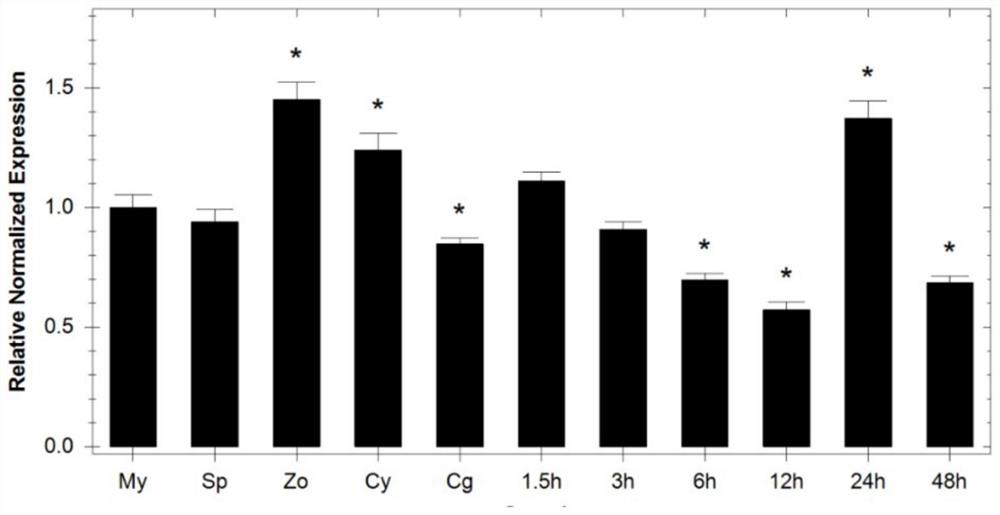
[0066] Example 2, Analysis of the expression pattern of PsZFAC gene in different developmental stages of Phytophthora soybean
[0067] Selected kit: Takara TB Green TMPremix Ex Taq TM II (Tli RNase H Plus) (Code: RR820A). Dilute 0, 4, 4 with mycelial stage cDNA samples 2 、4 3 、4 4 、4 5 、4 6 times, using PsActin (see SEQ ID No.13 and SEQ ID No.14 for primer sequences) as an internal reference gene and PsZFAC as a detection gene, a real-time fluorescent quantitative PCR standard curve was established. The amplification efficiency of gene primers needs to be above 90%, and an appropriate diluted sample concentration is selected for the next test.
[0068] Samples diluted 4 times at different developmental stages and infection stages were used as templates, PsActin was used as an internal reference gene, and real-time fluorescence was performed using detection primers PsZFAC-qPCR F / PsZFAC-qPCR (see SEQ ID No.5 and SEQ ID No.6 for the sequence) Quantitative PCR detection. ...
Embodiment 3
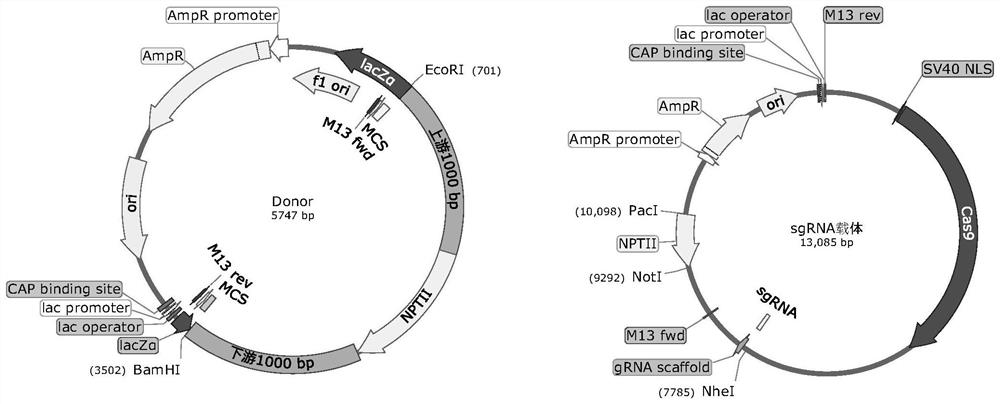
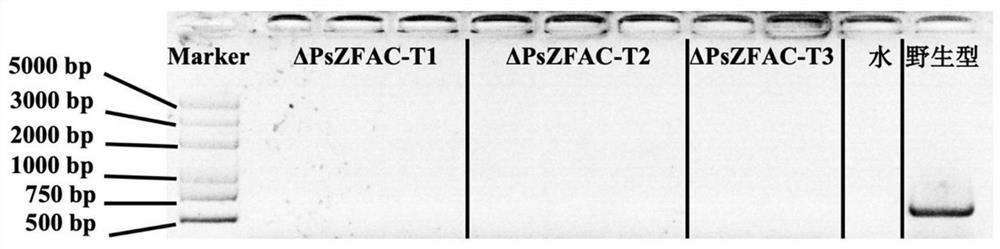
[0070] Embodiment 3, the acquisition of the knockout transformant of Phytophthora sojae PsZFAC gene
[0071] 3.1 Construction of the knockout vector of Phytophthora sojae PsZFAC gene
[0072] (1) Obtain candidate sgRNA
[0073] The sgRNA of PsZFAC was designed by EuPaGDT (http: / / grna.ctegd.uga.edu / ), and the full-length genome was searched on the website.
[0074] (2) Secondary structure screening
[0075] Use website tools
[0076] (http: / / rna.urmc.rochester.edu / RNAstructureWeb / Servers / Predict1 / Predict1.html) Analyze the secondary structure of candidate sgRNAs, select the number of hairpin complementary bridges that is less than or equal to 3 as candidate sgRNAs.
[0077] (3) Off-target analysis
[0078] The off-target analysis of sgRNA was carried out through FungiDB (www.fungidb.org), and a total of 23 bases including the PAM region were input for comparison, including 9-23 (including NGG) complete matches, and the risk of off-target was considered to be high.
[0079]...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com