Method of anticipating interaction between proteins
A prediction method and protein technology, applied in the field of prediction of protein-protein interactions, can solve problems such as lack of information on protein-protein interactions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
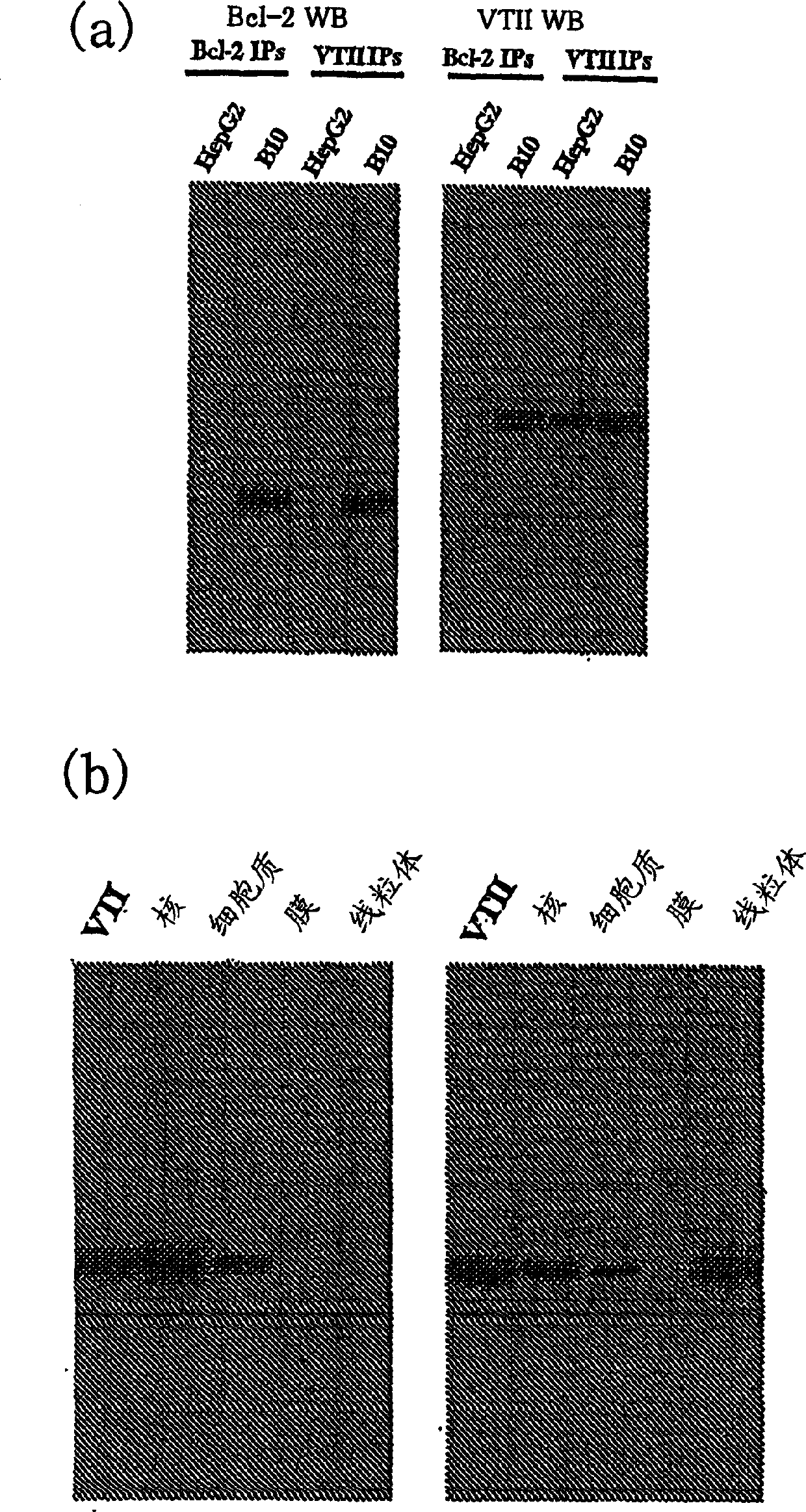
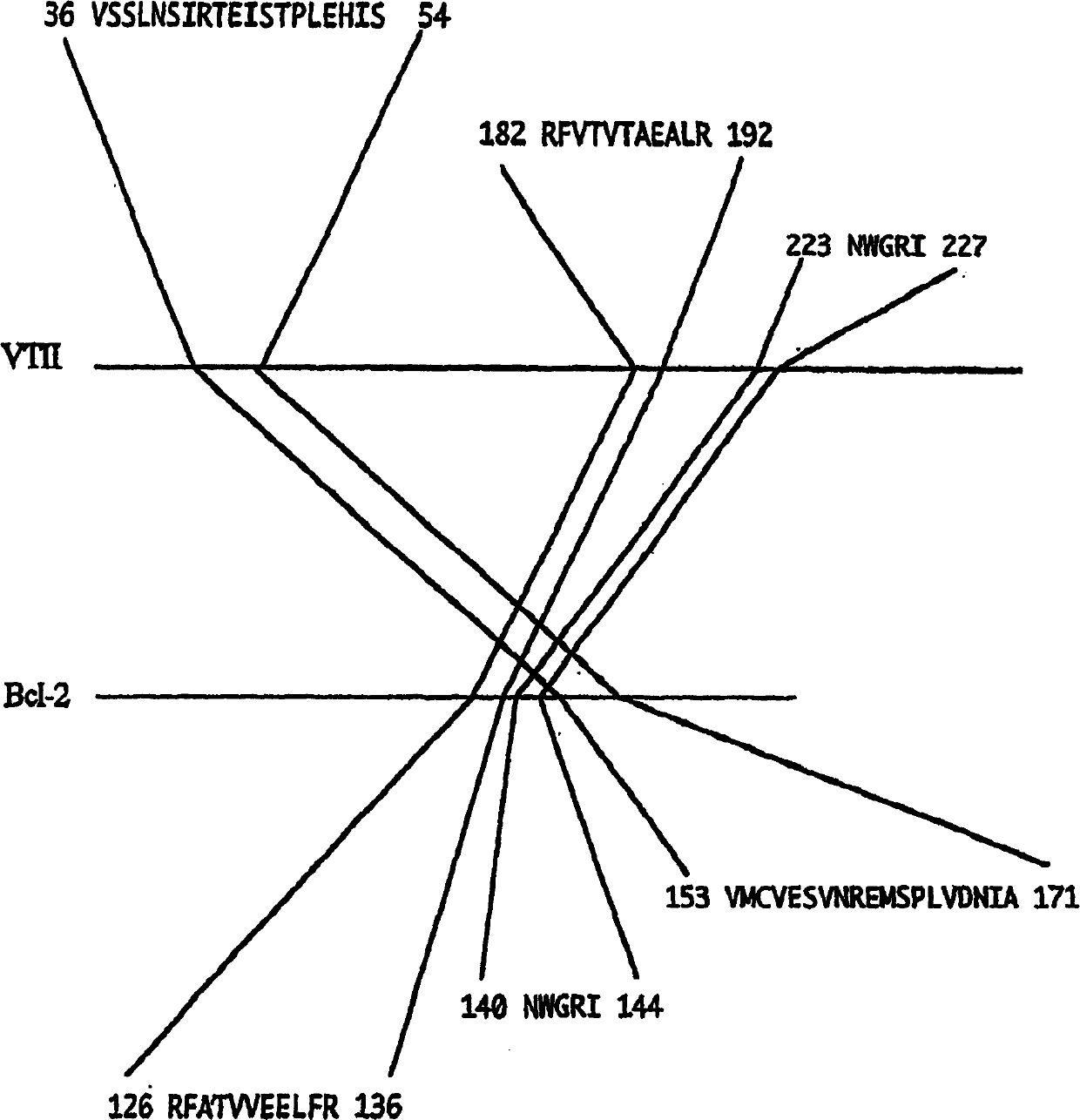
[0122] Fig. 1 is as the embodiment of step (1), is the first 20 residues [Fig. Oligopeptides [Fig. 1(b)]. Fig. 1(c) is an oligopeptide whose amino acid sequence is 6 long by decomposing the first 20 residues from the amino terminal of veirotoxin 2 (VTII). (Example 2)
Embodiment 2
[0123] In the step (4) of the method for predicting the interaction between proteins, the value calculated from the amino acid frequency and oligopeptide frequency obtained from the protein or polypeptide database is used as the predictive index of the interaction between proteins or polypeptides. The specificity of oligopeptides, calculated from the amino acid frequencies in proteins encoded by the E. coli genome shown in FIG. 4, is presented here as an example. The composition ratio Ai of each amino acid ai obtained from the frequencies of 20 amino acids in the total protein encoded by Escherichia coli [Fig. 4(a)] is shown in Fig. 4(b).
[0124] When the oligopeptide for specificity is a1a2a3a4a5, the specificity of the oligopeptide is calculated as A1×A2×A3×A4×A5. For example, when the oligopeptide is KCILF, the calculation is 4.406610×1.170608×6.004305×10.639652×3.898962×10 -10 . In addition, the specificity of oligopeptide LLLLL was calculated to be 136344.34×10 -10 , ...
Embodiment 3
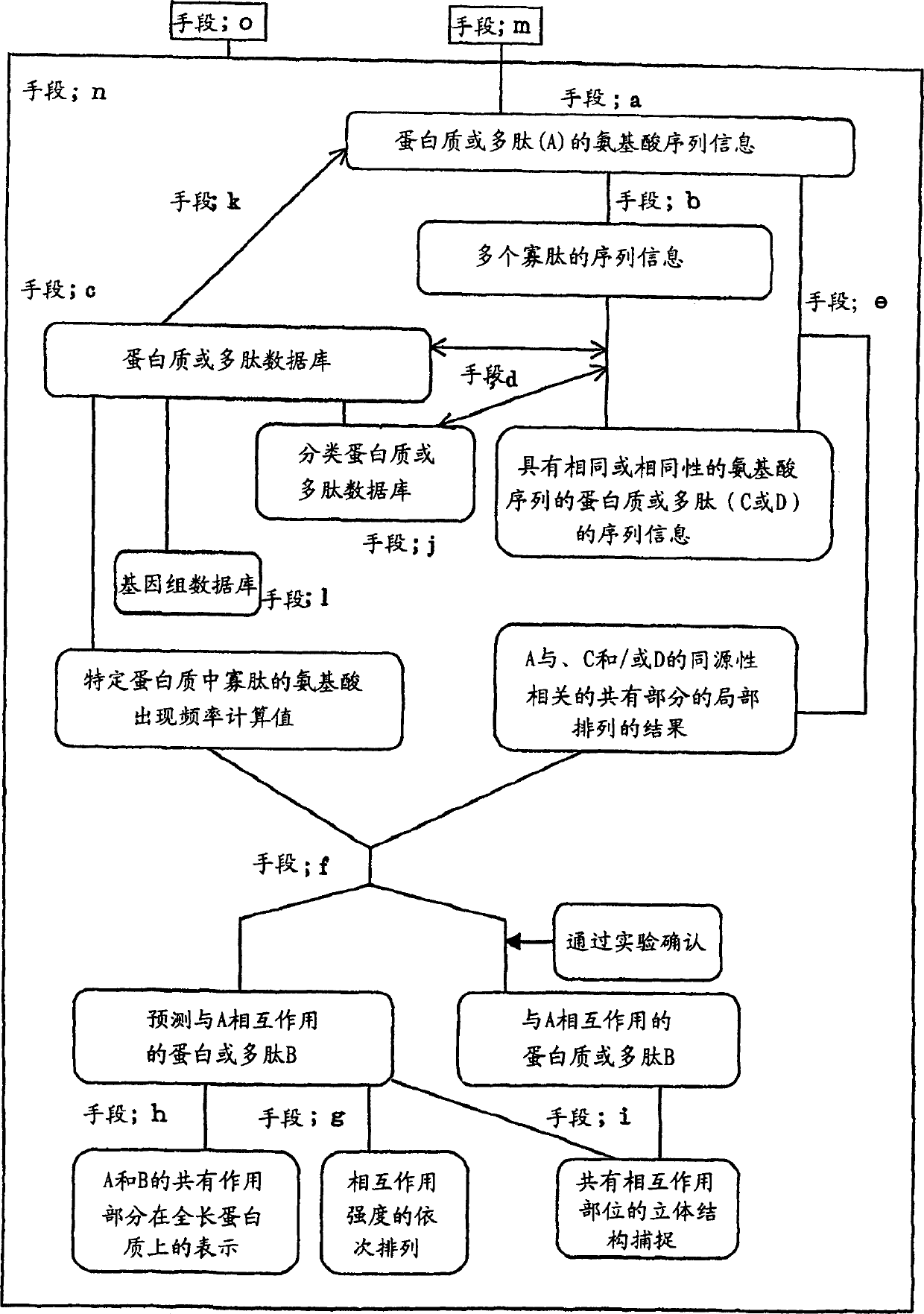
[0127] In the step (4) of the above-mentioned protein-protein interaction prediction method, the result of the local arrangement is used as an index for predicting the interaction between proteins or polypeptides, but here as an example, the method using Gotoh (Gotoh, O., Pattern matching of biological sequences with limited storage, Comput.Appl.Biosci.3, 17-20, 1987) for fractional case of partial sequence alignment. In the following example, when the score reaches 25.0 or more, the amino acid partial sequences between the protein or polypeptide (A) and the protein or polypeptide (B) are aligned (identical).
[0128] The m amino acid partial sequences between the protein or polypeptide (A) and the protein or polypeptide (B) are arranged, the score is Si (1≤i≤m), and the length of the amino acid sequence of B is LB. The index for predicting the interaction between A and B calculated from the results of this local arrangement is defined as sum(Si) / LB. The higher the index, the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com