Screening assays
A measurement method and selected technology, applied in the field of RNA biology, capable of solving problems such as complex calculations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0171] Embodiment A-experimental scheme:
[0172] a) Preparation of fluorescently labeled RNA. Using 5'O-dimethoxytrityl-2'O-triisopropoxymethyl-protected β'-cyanoethyl-(N,N-diisopropyl-)nucleotide Phosphoramides (GlenResearch), using published methods (see for example Chaix C. et al., Nucleic Acids Symp. Ser. 1989, (21): 45-6; Scaringe S.A. et al., Nucleic Acids Res. 1990, 18(18) : 5433-41) and the manufacturer's instructions, the 5' amino-C6 modified RNA was synthesized on a 394A synthesizer (Applied Biosystems). ORN was cleaved from the support, base-, phosphate- and 2'-deprotected, and purified by denaturing polyacrylamide gel electrophoresis according to standard methods. RNA concentration was calculated using UV-absorbance at 260 nm according to the Bouguer-Lambert-Beer Law; where used according to reference (Gray D.M. et al., Methods in Enzymology 1995, 246: 19-34) Determine the exact molar extinction coefficient at 260 nm. According to analytical RP-HPLC analysis (...
Embodiment B
[0189] Example B: Calculation method
[0190] Calculation of RNA accessibility P* using a constrained partition function M . Here we describe the P* of the secondary structure elements M For calculations of , the secondary structure elements are defined by sequence patterns with secondary structure constraints, such as NNUUNNUUU in a fully single-stranded conformation. P* M can be directly calculated as in equation [III] with respect to the setting of P*. In order to avoid numerical errors, P(α) is not calculated as a fraction of the partition function, but due to the difference in ensemble free energy, P(α)=exp((W-W α ) / RT), where W is the ensemble free energy of the RNA sequence, W αis the ensemble free energy of a structurally constrained thermodynamic secondary structure ensemble, where all sites in α form specific secondary structure elements. Ensemble free energy can be calculated using standard RNA secondary structure prediction software (Zuker M. Curr. Opin. Stru...
Embodiment 1
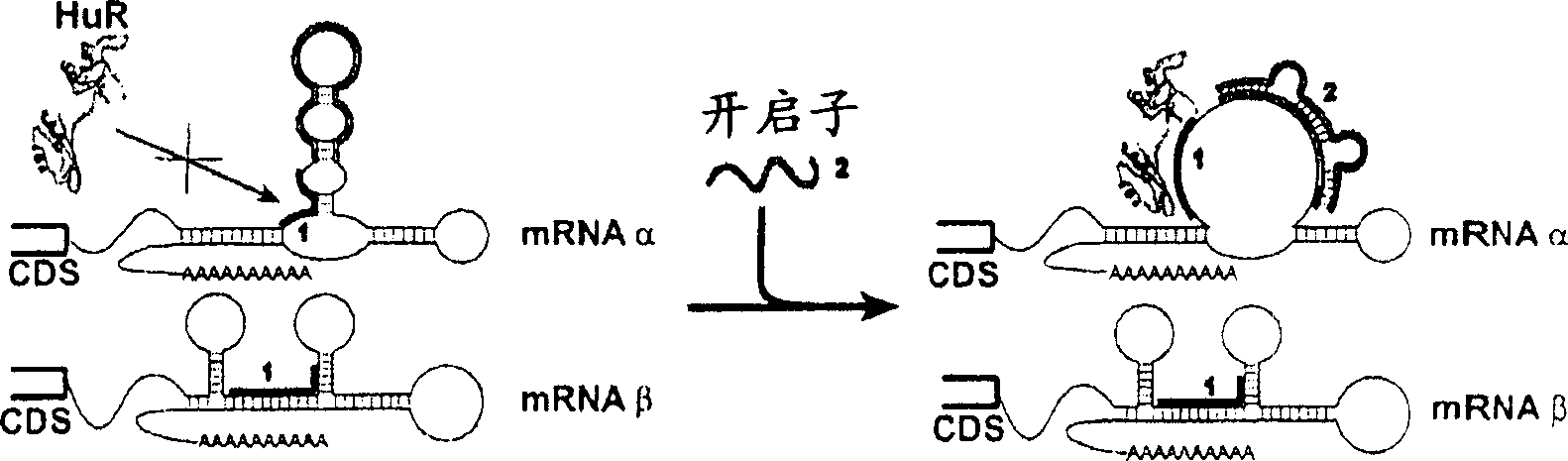
[0196] Example 1: Manipulation of HuR association with IL-2 mRNA, thereby manipulating IL-2 mRNA stability
[0197] Construction of opener, closer and negative control ORNs targeting IL-2 and TNF-α mRNA; experimentally validated in vitro and in human primary cell lysates.
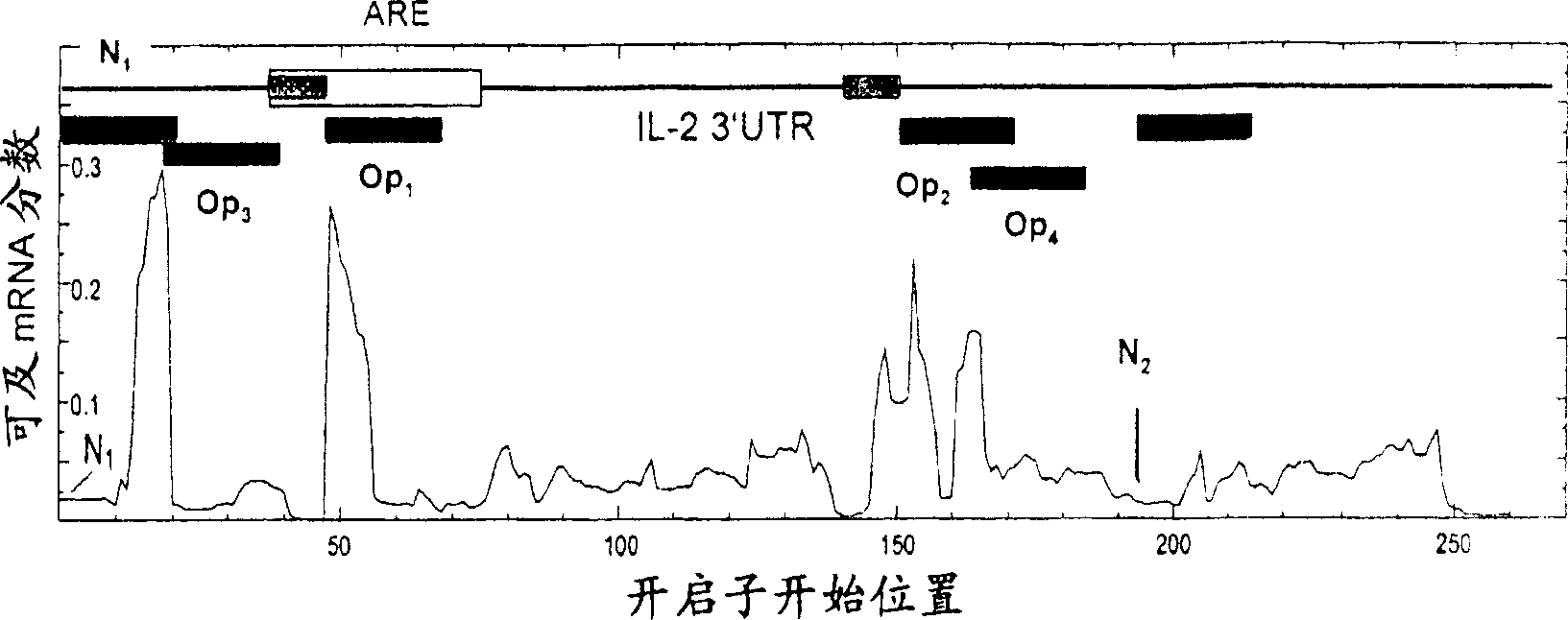
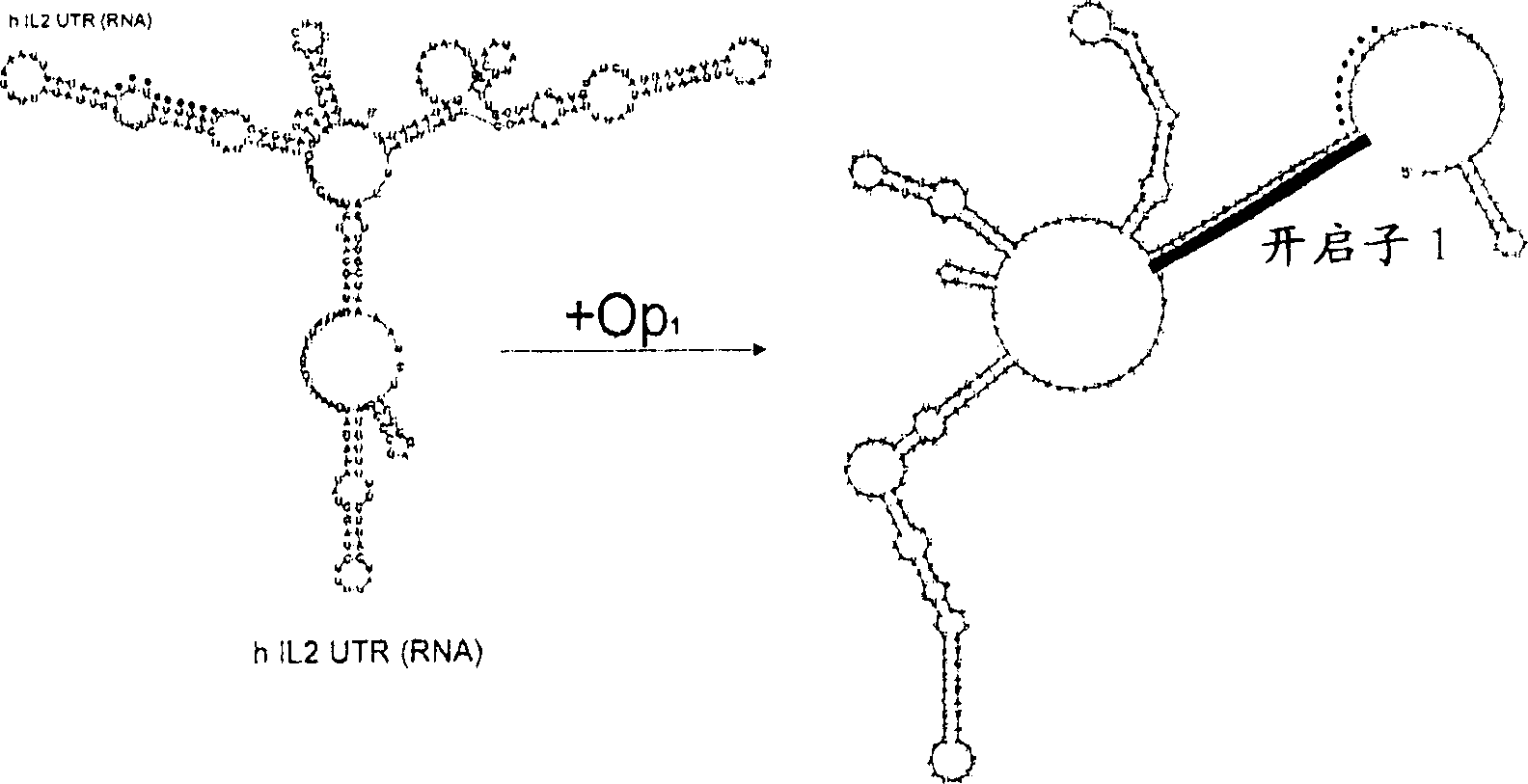
[0198] figure 2 Plots are presented showing the effect of hybridization of 20mer ORNs on the fraction of accessible RNA structures calculated for the purpose of designing openers and negative control ORNs directed against IL-2. Apparently, the dramatic increase in accessibility is restricted to a limited number of "hotspots" within the mRNA, mostly located externally and not near the HuR binding site. At many other positions, hybridization leaves the local ARE conformation barely affected. From these hotspots, 4 possible opener ORNs and 2 negative control ORNs were selected for experimental characterization (Table 1). In the first step, we used ss ORN.
[0199] Opener roles were initially identified in...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com