MDM2-Containing Double Minute Chromosomes And Methods Therefore
a technology of double-minute chromosomes and mdm2, applied in the field of molecular diagnostics, can solve the problems of difficult to use the output of currently known methods to reassemble large regions of tumor genomes, require significant enrichment efforts that are only feasible, and unsuitable for initial tumor diagnosis, so as to achieve rapid and comprehensive identification of genetic rearrangements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
examples
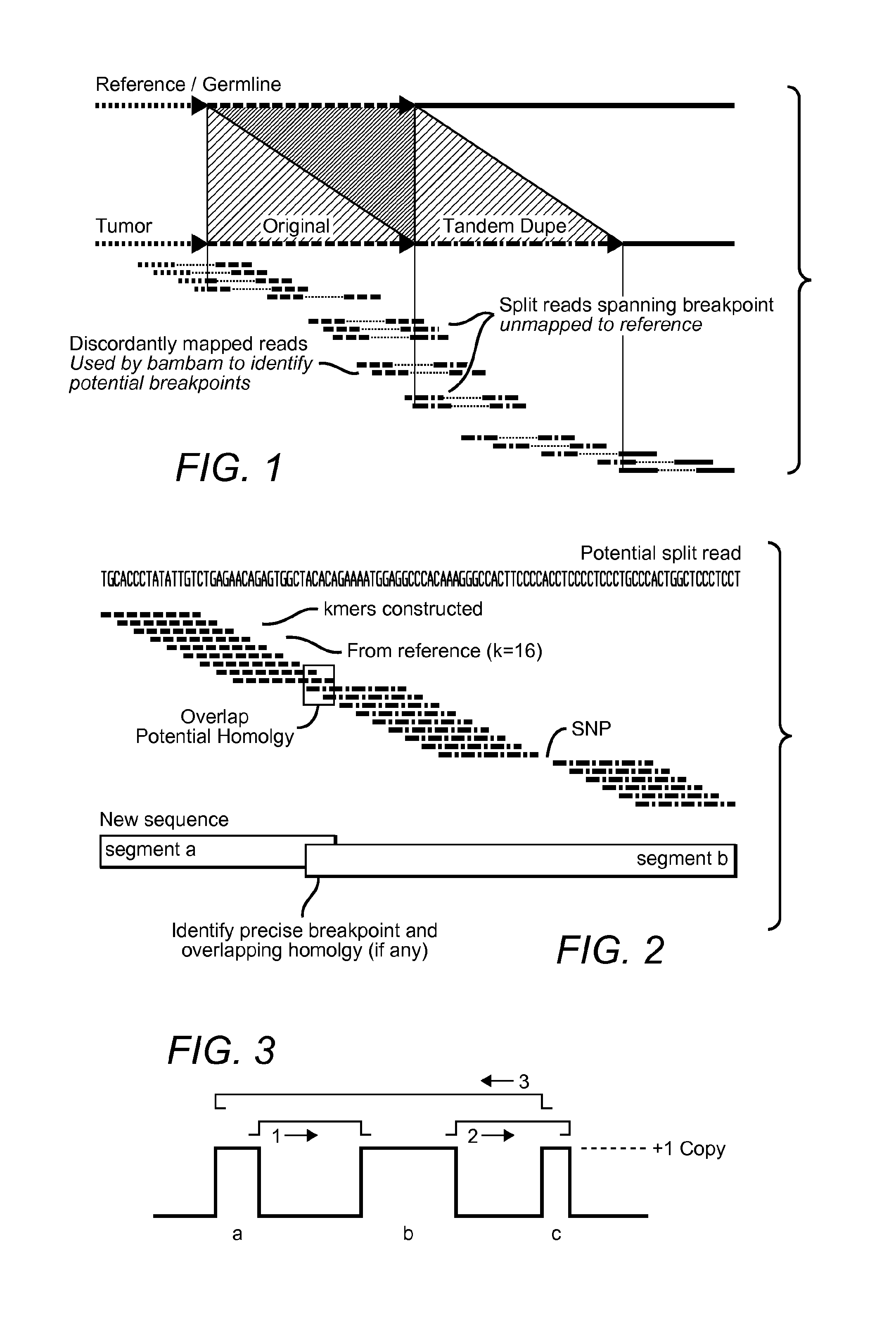
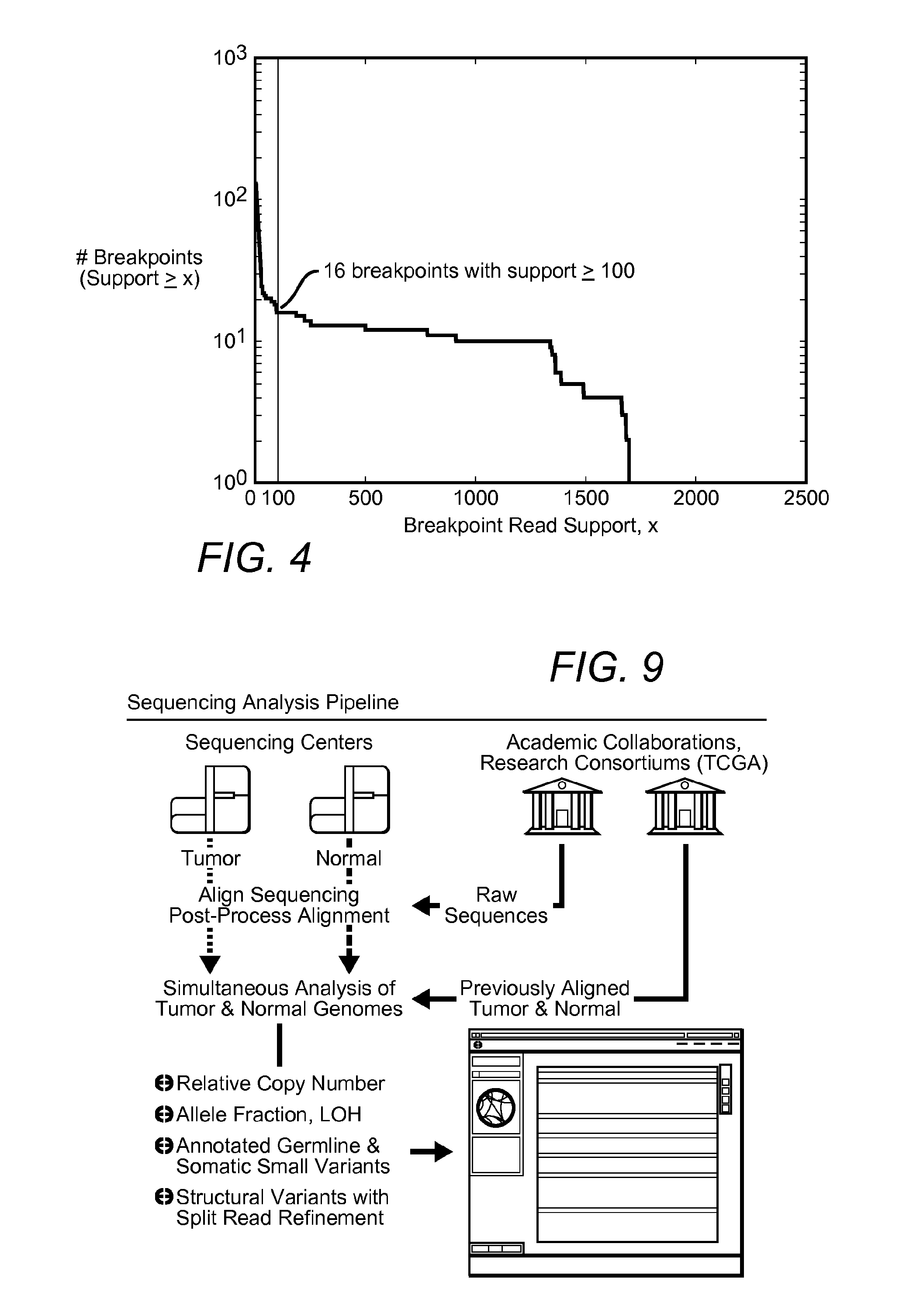
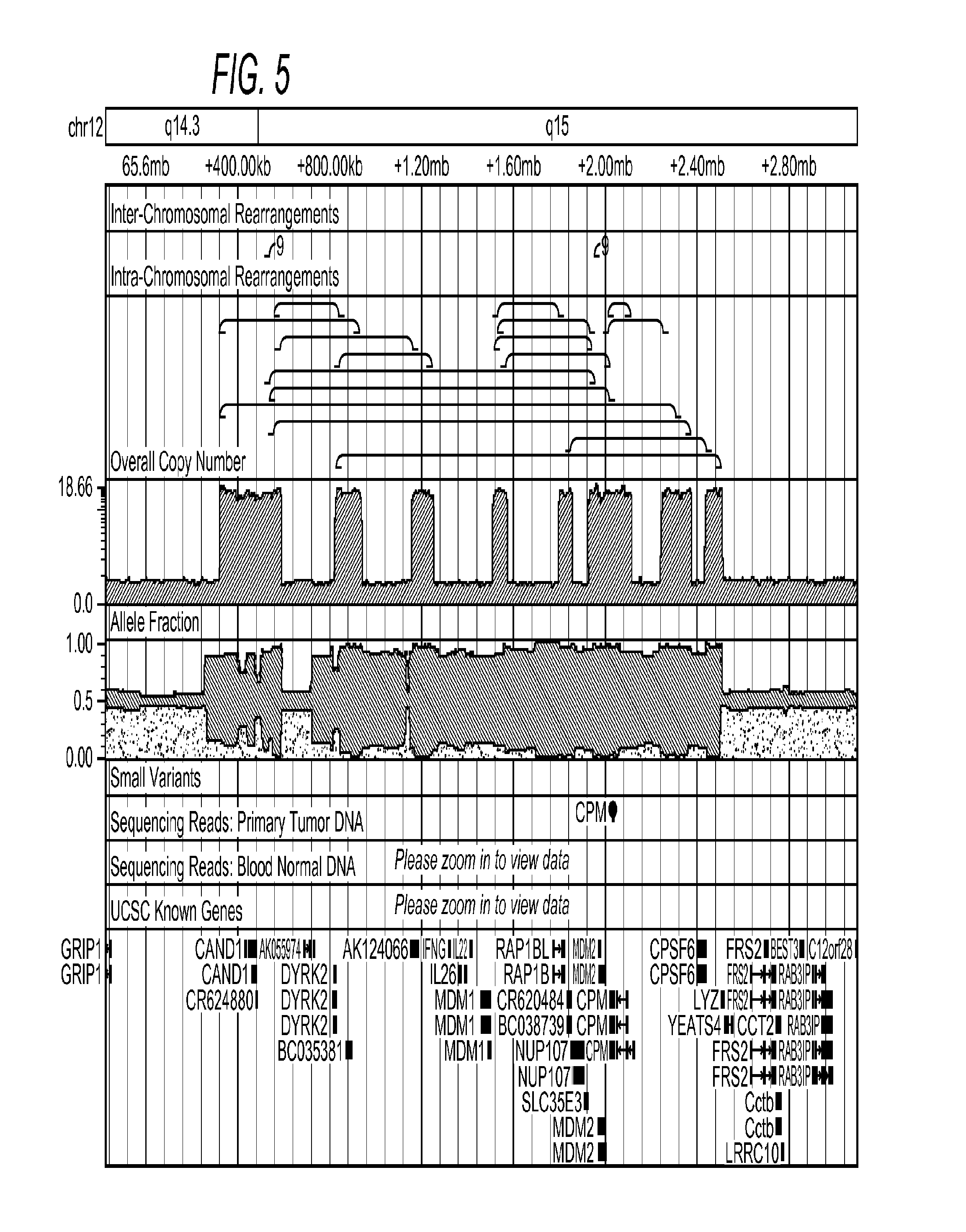
[0045]The inventors applied the above described methods to two glioblastoma multiforme (GBM) samples designated TCGA-06-0648 and TCGA-06-0152, both sequenced by The Cancer Genome Atlas (TCGA) project. The tumor and matched normal (blood) sequencing datasets from these samples were processed as described in Methods, producing tumor vs. normal relative copy number estimates, identifying breakpoints, and performing split read analysis. The tumor and normal genomes of both samples were sequenced to an average coverage of approximately 30×.
[0046]A total of 3,696 breakpoints were identified by bambam, of which 132 breakpoints were found by bridget to have split reads directly spanning the putative breakpoint. FIG. 4 shows a histogram of read support for all somatic breakpoints found in sample TCGA-06-0648. By setting the minimum read support threshold to 100, all but 16 of the somatic breakpoints supported by bridget were removed. Interestingly, all 16 of these highly supported breakpoint...
PUM
| Property | Measurement | Unit |
|---|---|---|
| read support threshold | aaaaa | aaaaa |
| structure | aaaaa | aaaaa |
| resistance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com