Engineering t cell receptors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Selection of a Model TCR
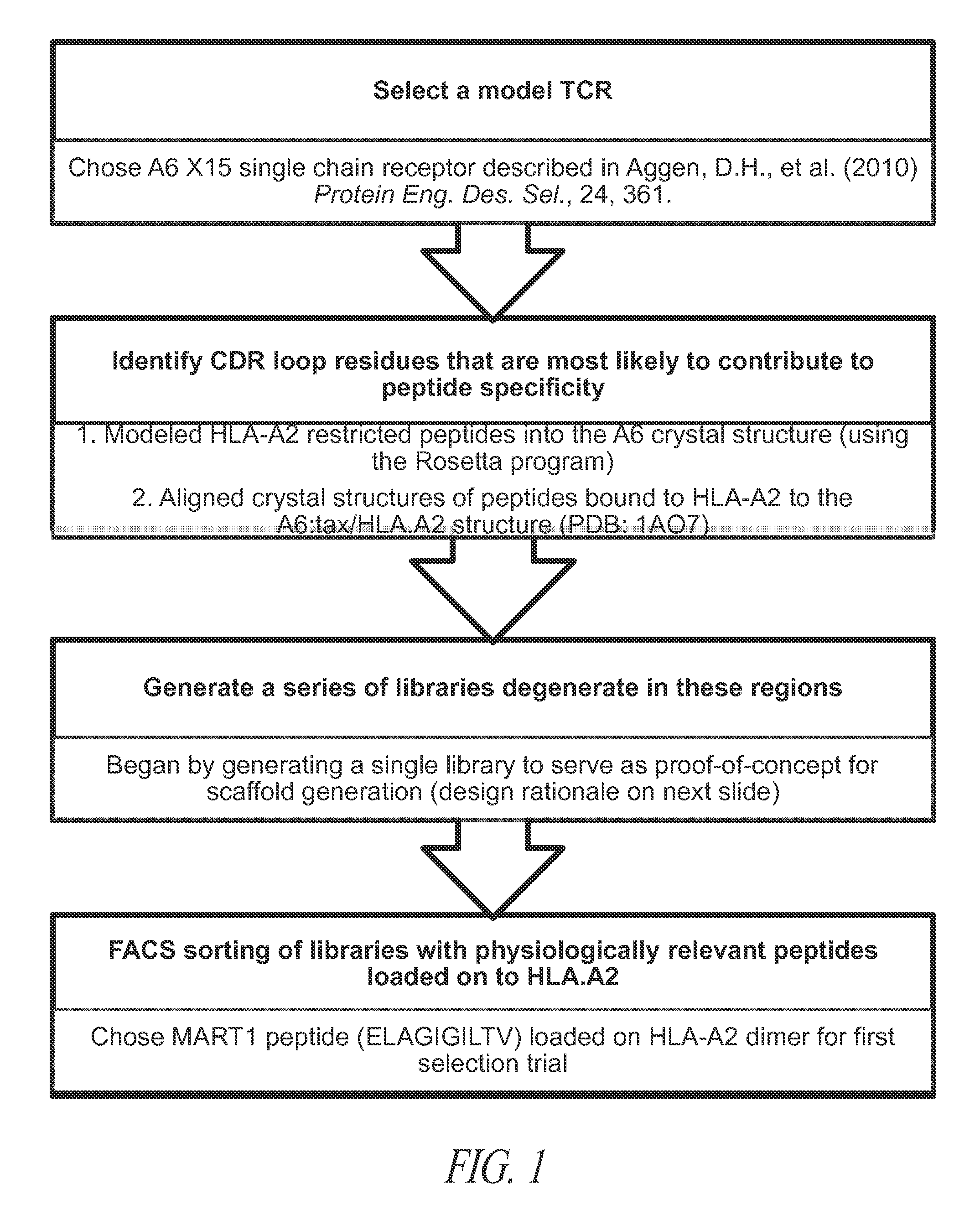
[0237]TCRs all adopt a similar Ig-fold and docking angle, and TCR recognition of pepMHC is mediated entirely by specific residues on CDR loops (Garcia et al. (2009) Nat Immunol, 10, 143-7; Marrack et al. (2008) Annu Rev Immunol, 26, 171-203; Rudolph et al. (2006) Annu Rev Immunol, 24, 419-66)). Hence, according to the present invention, a single TCR with known structure provides a scaffold for in vitro engineering with specificity and high affinity against non-cognate peptides displayed on MHC. That is, by generating mutants with degenerate residues within CDR loop residues that are most likely to directly contact peptide, libraries of mutants within a single TCR were generated in order to provide TCRs that can be developed having high affinity against non-cognate peptide-MHC antigens.
[0238]The general strategy used to discover, or generate, novel TCRs against non-cognate antigens from a single scaffold is shown in FIG. 1. The process involves: selecting a si...
example 2
Analysis of the Human TCR A6 in Complex with Tax:HLA.A2 as a Scaffold for TCR Engineering
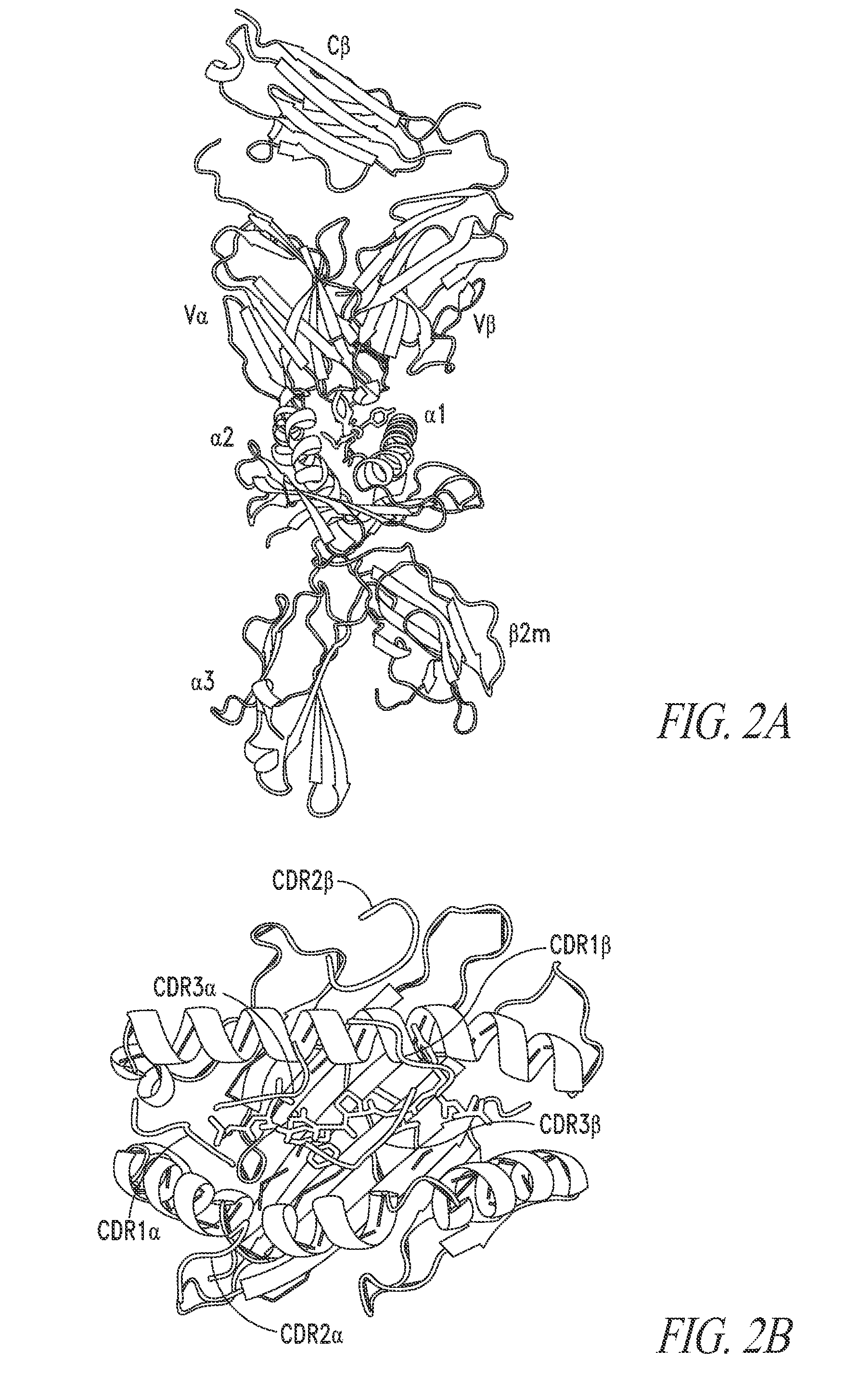
[0242]For illustrative purposes, the A6 TCR was selected as the single TCR having a known structure. The structure of the A6:Tax peptide:HLA-A2 complex (PDB: 1AO7) (Garboczi et al. (1996) Nature, 384, 134-141), was published in 1996. The side view of the complex showed that the ends of the variable domains that contained the six CDRs docked onto the Tax:HLA.A2 molecule, with the central region of the binding site positioned over the peptide Tax (FIG. 2A) The top down view of the Tax:HLA.A2 complex, with the TCR “removed”, except for the six CDR loops. This view shows that the TCR adopts a diagonal position over the peptide-MHC, a finding which has now been observed for all TCR:peptide-MHC structures. In this orientation, the two CDR3 loops are positioned over the peptide, while there are various residues from CDR1 and CDR2 loops that interact predominantly with the helices of the MHC molecule. T...
example 3
Analysis of the CDR Loop Residues Most Likely to Contribute to Peptide Binding and Specificity
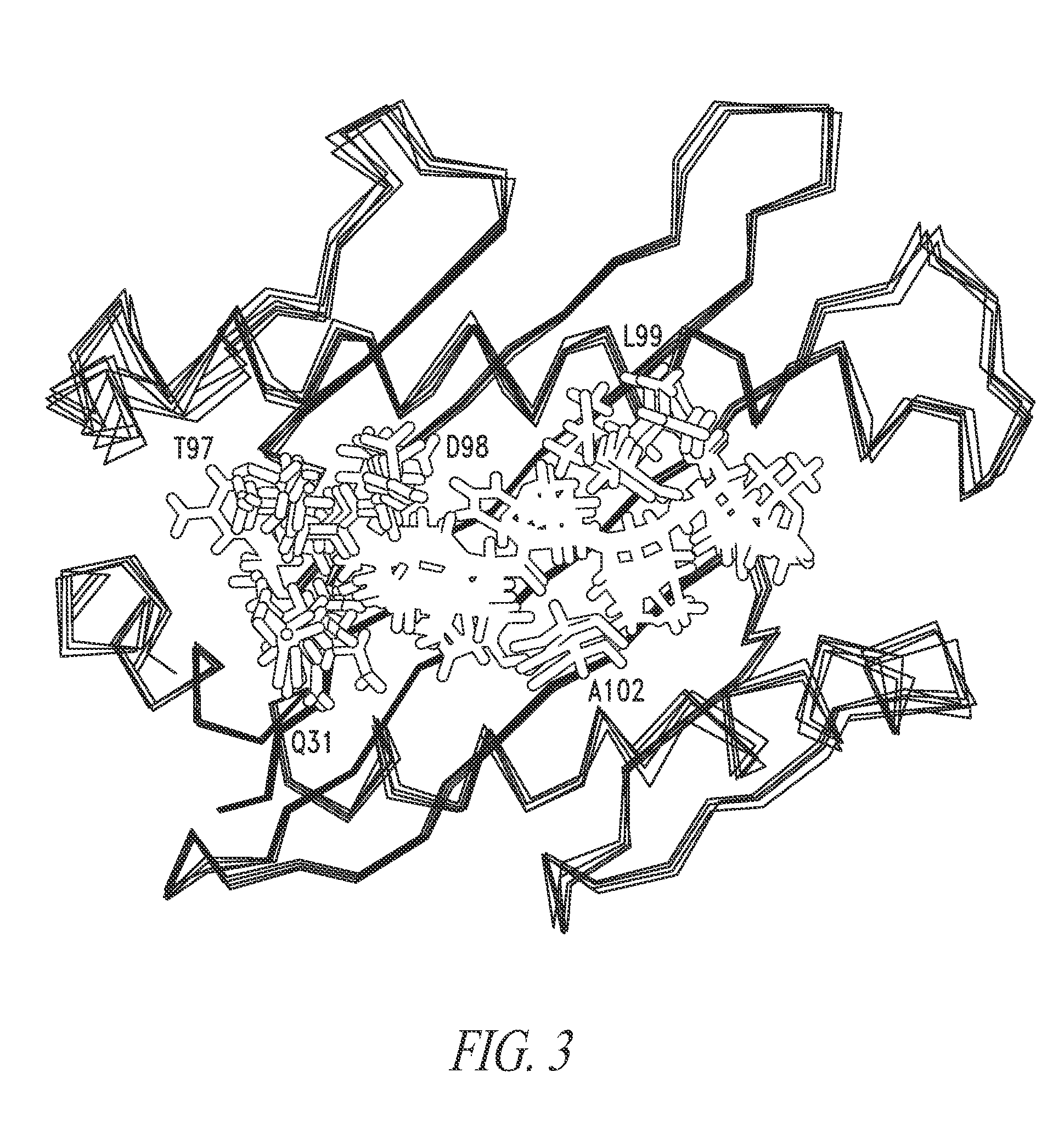
[0244]In order to identify potential contact and specificity-determining residues, various approaches were used to determine which residues of the A6 CDR loops would be most likely to accommodate, and provide binding energy to, a wide array of peptides in the HLA.A2 peptide-binding groove. First, a panel of other HLA.A2 restricted peptides was modeled into the A6 crystal structure (FIG. 3). Using the A6:Tax peptide:HLA.A2 crystal structure (PDB:1AO7) as a starting point (Garboczi et al. (1996) Nature, 384, 134-141), the Rosetta Backrub modeling program was used to model the HLA.A2 restricted peptides (i.e., Tax, Mart1-9 mer, Mart1-10 mer, SL9 HIV, WT1, and Survivin) into the HLA.A2 groove using Rosetta Backrub flexible backbone modeling algorithms (FIG. 3) ((Lauck et al. (2010) Nucleic Acids Res, 38, W569-75); kortemmelab.ucsf.edu / backrub / ). The peptides that were modeled into the binding g...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
| affinity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com