Signatures and determinants for distinguishing between a bacterial and viral infection and methods of use thereof
a technology of determinants and signs, applied in the field of identification of biological signatures and determinants associated, can solve the problems of limiting the effectiveness of current diagnostic solutions for reducing, ineffectiveness and inappropriateness, and current solutions (such as culture, pcr and immunoassays) do not meet all these requirements, so as to prevent unnecessary antibiotic treatment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
ethods
[0317]In-Vivo Clinical Study Protocol
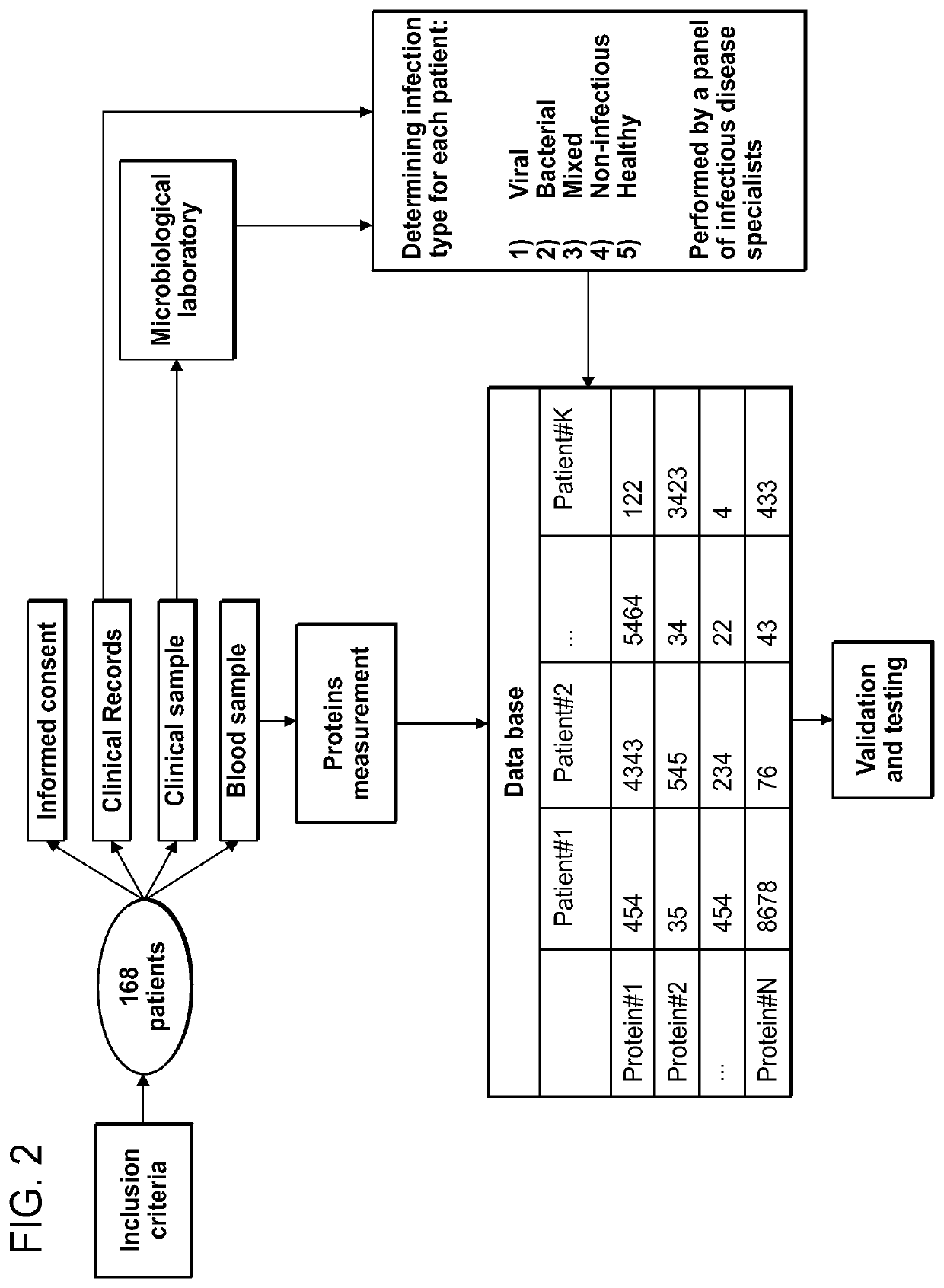
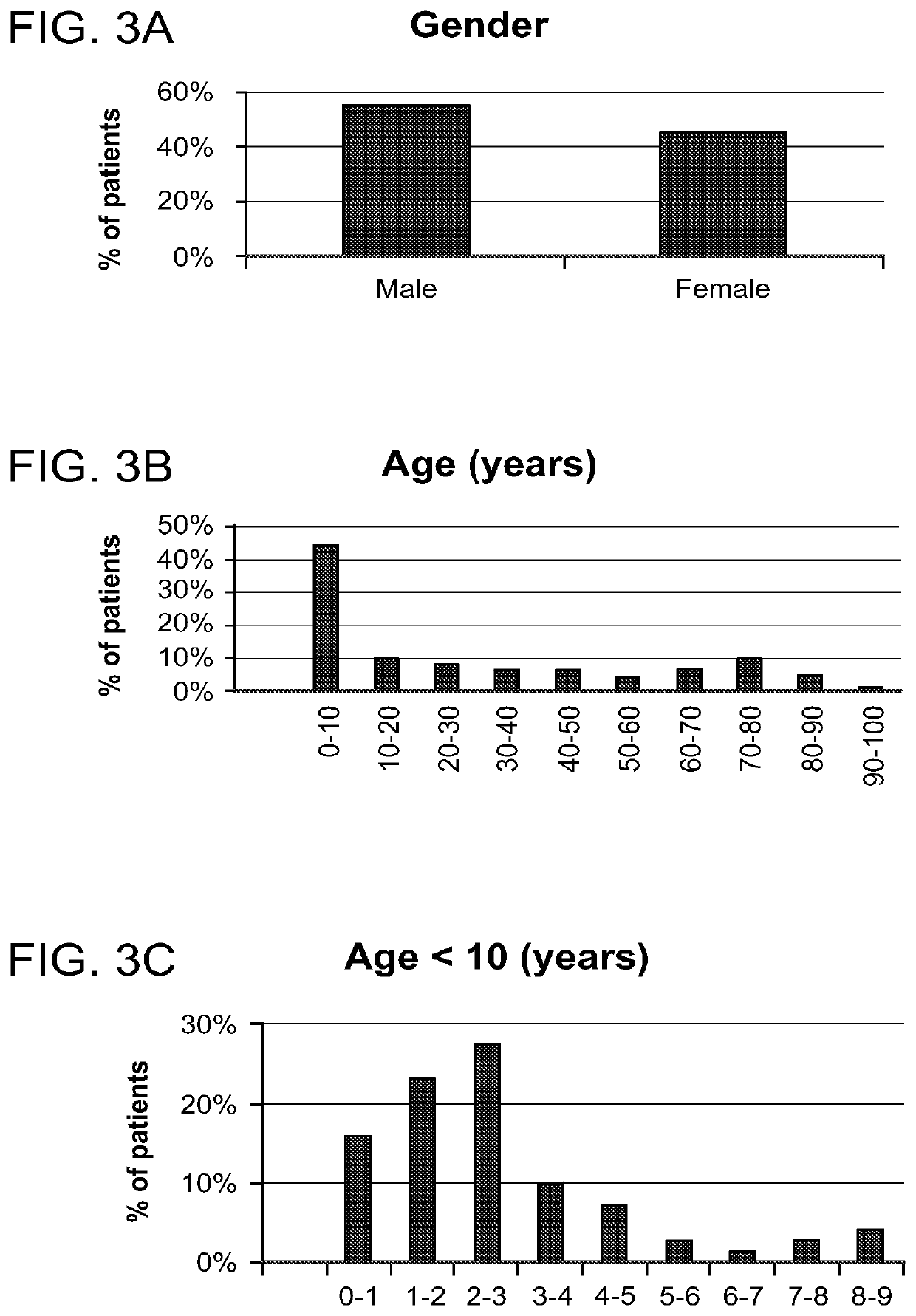
[0318]To screen for potential DETERMINANTS that can separate between different sources of infection we performed a clinical study on 168 hospital patients. Study workflow is depicted in FIG. 2. Briefly, after signing an informed consent, 2-6 ml of peripheral venous blood was collected in EDTA containing CBC tubes. The blood was than stored in 4 degrees Celsius for 1-4 hours. Depending on the type of infection, patient specimens were also collected (e.g. nasal swab and urine) for microbiological analysis. The clinical history, the clinical lab results, the imaging data and the microbiological results are used to identify the source of infection by an infectious disease expert. DETERMINANT measurements and infection source related data were sotred in a database that was used to screen and then test the differential accuracy of each individual DETERMINANT and the multi-DETERMINANT signature.
[0319]Patient Inclusion & Exclusion Criteria
[0320]Pat...
example 2
rminant Polypeptides are not Differentially Expressed in Different Types of Infections
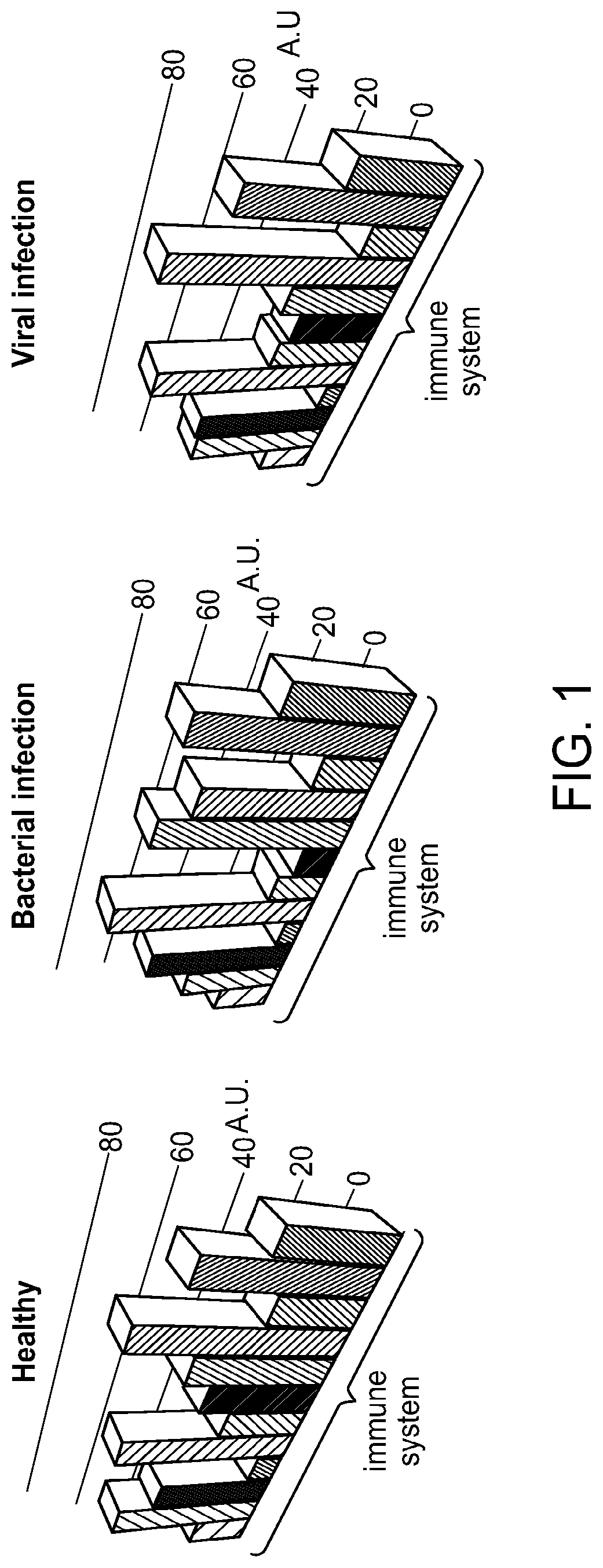
[0414]We have determined that most DETERMINANT polypeptides are not differentially expressed in patients with different types of infections. Moreover, DETERMINAT-polypeptides that have a well-established mechanistic role in the immune defense against infections are often not differentially expressed. Thus an immunological role of DETERMINAT-polypeptides does not necessarily imply diagnostic utility. This point is illustrated in FIG. 5, which shows examples of DETERMINANT polypeptides with an active immunological role in the host response to bacterial or viral infections. We find that the levels of these DETERMINANT polypeptides were not significantly differentially expressed in lymphocytes of patients with viral versus bacterial infections.
example 3
nts that Differentiate Between Bacterial Versus Viral Infected Patients
[0415]We identified a few DETERMINANTS that were differentially expressed in patients with bacterial versus viral infections in a statistically significant manner (Wilcoxon ranksum P<0.001). DETERMINANT names and classification accuracies are listed in Table 1A. The distributions and individual patient measurements for each of the DETERMINANTS are depicted in FIG. 4A (dots corresponds to DETERMINANTS measurement in individual patients and bars indicate group means). The abbreviations lym, gran, mono, mean and total are used to indicate whether a DETERMINANT polypeptide was differentially expressed in lymphocytes, granulocytes, monocytes, mean signal over all leukocytes or total signal of leukocytes respectively. The latter was measured in a constant volume of 1 ul of blood. Each subplot corresponds to a different DETERMINANT.
PUM
| Property | Measurement | Unit |
|---|---|---|
| electrophoretic detection | aaaaa | aaaaa |
| time | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com