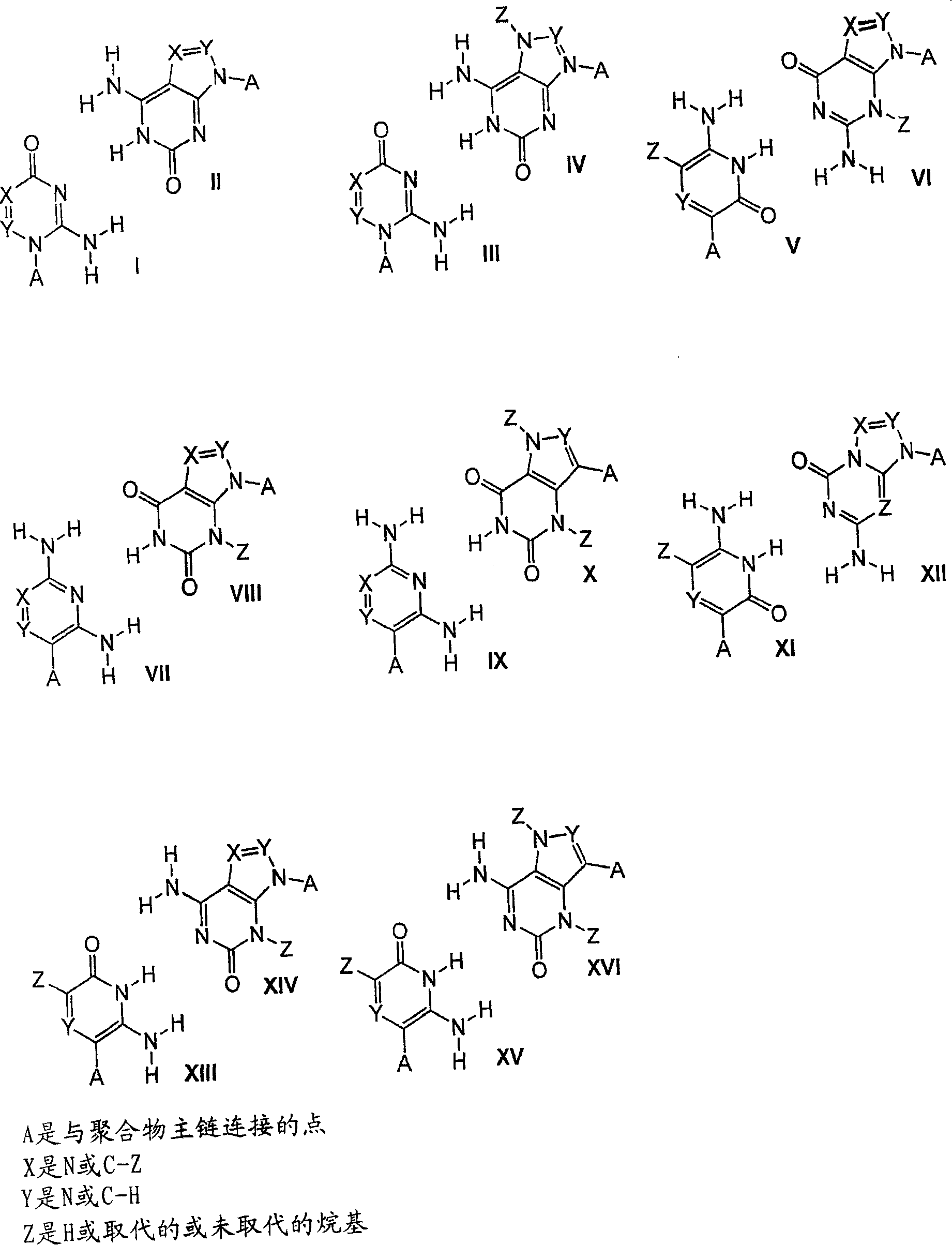
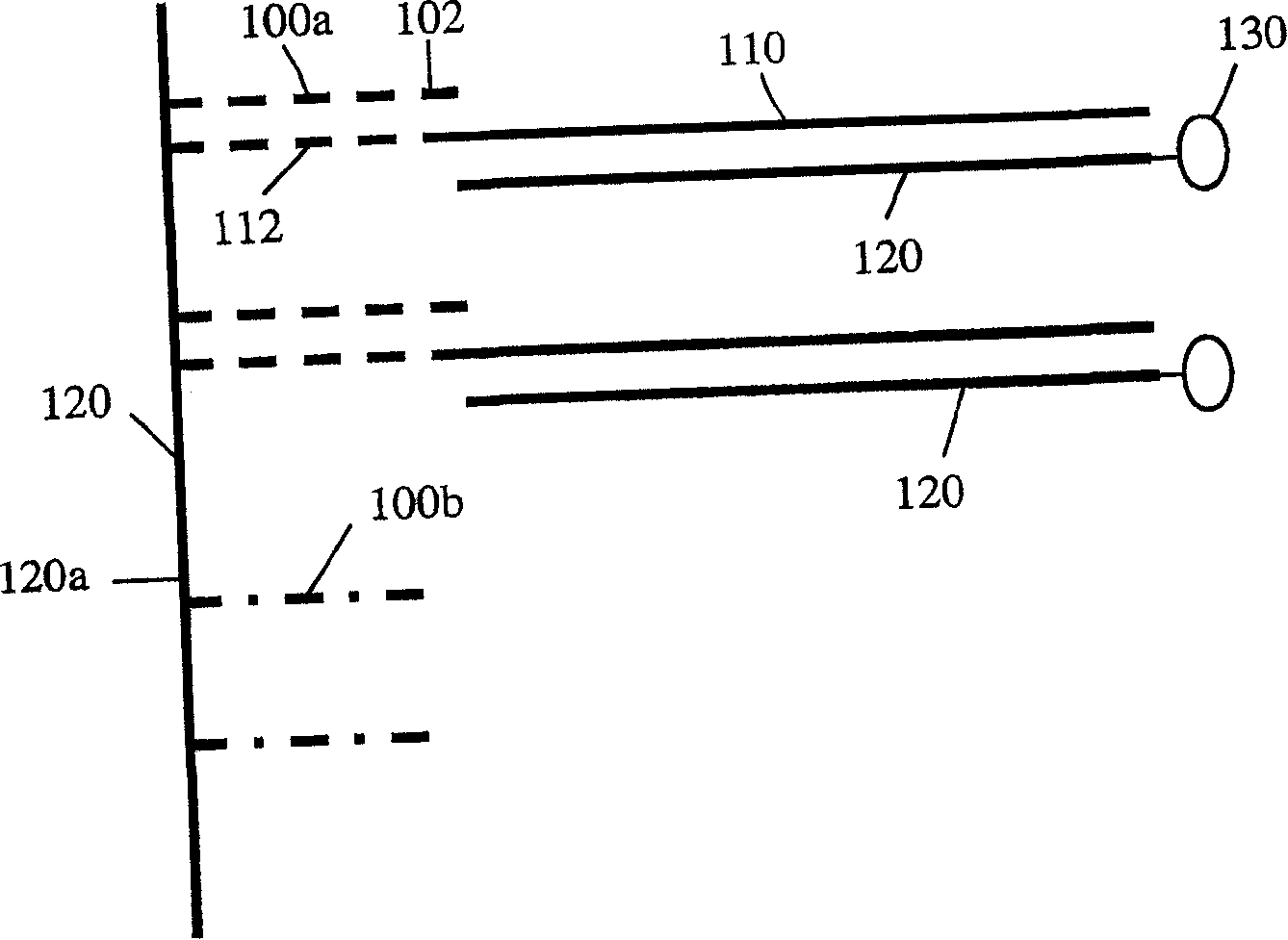
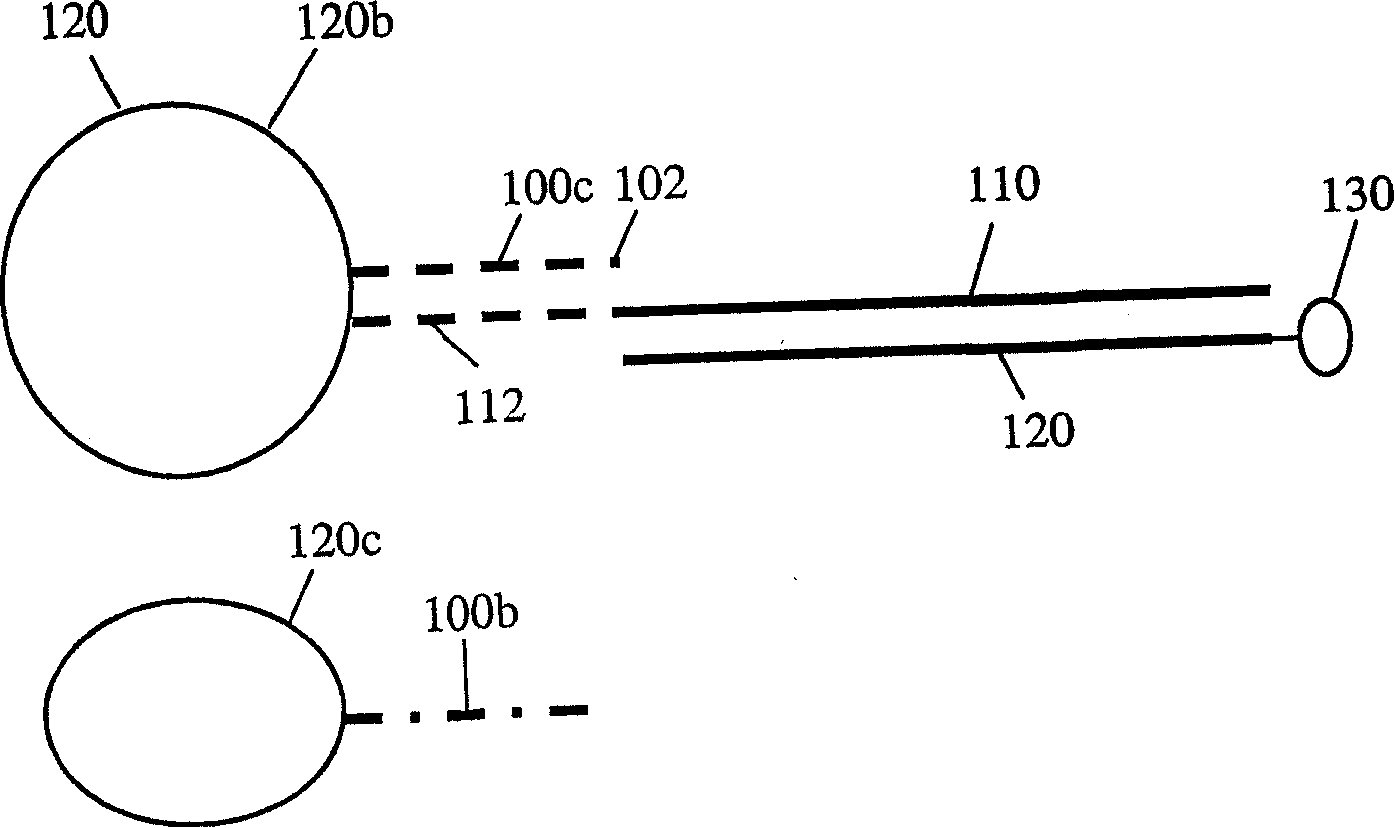
Solid support assay systems and methods utilizing non-standard bases
A non-standard, base technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as increased non-specific hybridization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0149] Cross-hybridization detection of coding and marker sequences
[0150] Instruments used in this analysis included the Luminex 100 and Luminex Microbeads, DNA synthesizer (Northern Engineering Company), spectrophotometer for point measurement synthesis field, thin-layer chromatography column (TLC) (SI250F TLC silica gel plate, JTBaker), qualitative control for oligonucleotides, centrifuge , Sonicator (Ney Dental), Vortex (Vortex) and various pipettes (2, 20, 200 and 1000 μl)
[0151] A set of at least 100 oligonucleotide strands (molecular recognition sequences) and their complements (tag sequences) are designed and synthesized. These two sets of oligonucleotides contain non-standard nucleotides (isoC and isoG) (EraGen Biosciences, Inc., Madison, WI) and natural nucleotides (A, G, C and T) (Perkin-Elmer / ABI) , 9 to 10 bp long. The 5' end of the first set of oligonucleotide strands designed as molecular recognition sequences was labeled with an amino modifier (C6-TFA...
Embodiment 2
[0170] Preliminary determination of the role of non-standard bases in predicting nucleic acid duplex stability parameters
[0171] This experiment uses a Beckman DU-7500 spectrometer with temperature control system and sample holder. The precise temperature control system can measure the temperature of six samples at the same time. The quartz cups are Hellma products from the United States. In order to cover a range of one hundred times the sample concentration, the optical path lengths are 0.1cm, 0.2cm, 0.5cm, and 1.0cm. DNA was synthesized on a DNA synthesizer model 392 of Perkin-Elmer / ABI Company. Liquid tank (Fisher) for TLC chromatography, TLC plate (Si250F, JT Baker). Sanvant SpeedVac was used to prepare DNA, and Sep-pakC-18 purification column (Waters), UV lamp, vortex shaker, 100cc syringe, and several micropipettes (2, 20, 200, 1000 μl) were also used.
[0172] Oligonucleotides were synthesized using natural nucleotides (A, G, C and T) (Perkin-Elmer / ABI) and isoC...
Embodiment 3
[0197] Embodiment 3 and comparative examples thereof
[0198] site-controlled incorporation
[0199] First Primer 5' AGAACCCTTTTCCTCTTCC (SEQ ID NO: 104) Target Sequence 5' AAGAACCCTTTCCTCTCCGATGCAGGATACTTAACAATAAATATTT (SEQ ID NO: 105)
[0200] Second primer CTACGTCCTATGAATTGTTATTTATAAAYAGGACAGACG5' (SEQ ID NO: 106)
[0201] Y=isoCTP
[0202] The first primer and its target sequence and the second primer are shown in SEQ ID NO: 104, SEQ ID NO: 105, and SEQ ID NO: 106, respectively.
[0203] PCR experiments were performed with the following mixture: 0.2 μM first primer, 0.2 μ / M second primer, 50 fM target sequence, dGTP, dATP, dTTP and dCTP, 50 μM each, 10 mM TrispH8, 0.1% BSA, 0.1 % Triton X-100, 0.1 μg / μl degraded salmon sperm DNA, 40 mM KAc, 2 mM MgCl 2 , 1 U Amplitaq Stoffel (Perkin Elmer Biosciences, Foster City, CA). The mixture was first incubated at 95°C for 2 minutes, then cycled at 95°C for 1 second and 58°C for 10 seconds, and 30 PCR cycles were set. The final...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com