Malic dehydrogenase related to high photosynthetic capacity and resisting reversal of wheat, coded genes and method for breeding plants of resisting reversal
A technology of malate dehydrogenase and encoding, which is applied in botany equipment and methods, biochemical equipment and methods, genetic engineering, etc., and can solve the problems of low photosynthetic efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1. Isolation and sequence analysis of wheat malate dehydrogenase gene probe
[0029] Wheat (Triticum aestivum L) was planted in a greenhouse, watered and fertilized normally, until it grew to 2-3 internodes, and roots, stems, and leaf tissues were collected and tested with TRI reagent (Molecular Research Center, Inc, Cincinnati, USA) Total RNA was extracted with PolyAT mRNA Isolation kit (Promega company, Madison, USA) separates Poly(A) + RNA, the content and purity of RNA were analyzed by UV spectrophotometer.
[0030] The conditions for reverse transcription to synthesize the first strand of cDNA are: Poly(A) + RNA 1ug, 5μmol / L Primer 5'-GACTCGAGTCGACATCGA(T) 17 -3', 0.5mmol / L dNTP, 50 units of RNasin, incubate at 65°C for 10 minutes, after cooling on ice, add 200U SuperScript TM IIRNase H-Reverse Transcriptase (Gibco), incubated at 42°C for 1 hour, added EDTA to a final concentration of 10 mM, incubated at 95°C for 10 minutes, and cooled on ice.
[00...
Embodiment 2
[0034] Example 2. Screening and sequence analysis of wheat malate dehydrogenase gene
[0035] With Wheat Stem Poly(A) + Using RNA as a template, cDNA was synthesized with a ZAP vector (Stratagene, LaJolla, CA, USA), and packaged in vitro to construct a cDNA library of wheat stems. Take 50ng probe DNA, add 50uCi 32 P-dCTP was labeled, incubated at 37°C for 1 hour, and EDTA was added to a final concentration of 10mM to terminate the reaction. After the probe passed through the Sephadex G-50 column, it was boiled at 100° C. for 10 minutes, and immediately cooled on ice for hybridization.
[0036] Take 50000pfu to plate, and transfer it to nitrocellulose membrane, denature in 0.5M NaOH plus 1.5M NaCl for 2 minutes, 1.5M NaCl plus 0.5M Tris-HCl (pH8.0) for 5 minutes, 0.2M Tris- Rinse with HCl (pH7.5) plus 2 x SSC for 30 seconds, sandwich the membrane between two layers of Whatman filter paper, bake in vacuum at 80°C for 2 hours, then add hybridization solution (6 x SSC, 5 x Denh...
Embodiment 3
[0039] Embodiment three, the translation of wheat malate dehydrogenase gene coded protein
[0040] Artificially synthesize a pair of primers:
[0041] 5'-Primer: 5'-CGGGATCCATGGCTGCGAAGGAACCGATG-3'
[0042] 3'-primer:
[0043] 5'-ATAAGAATGCGGCCGCTTACGCCAGGCATGAGTAGG-3'
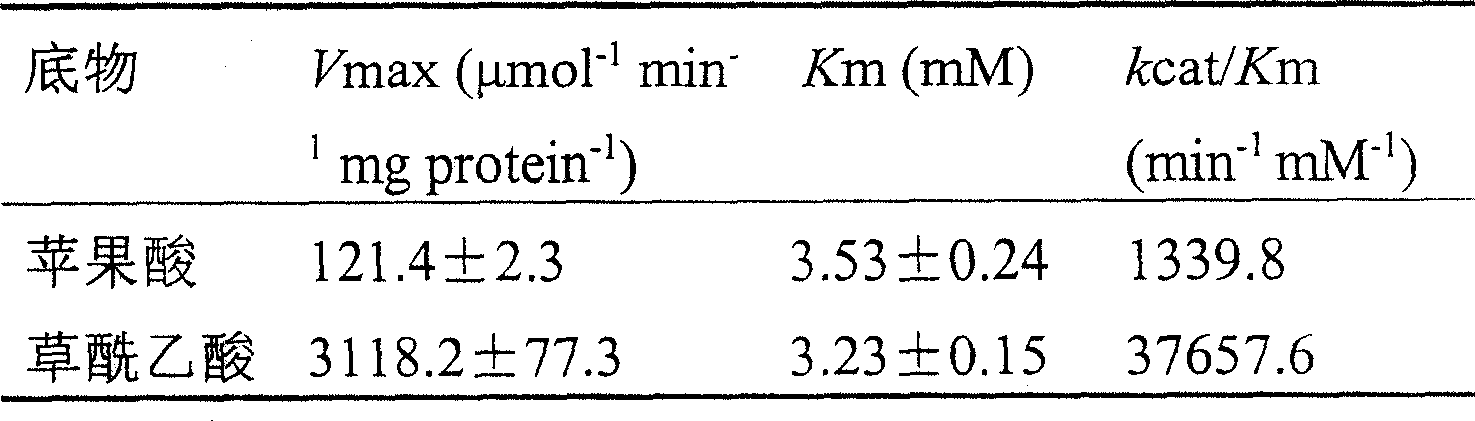
[0044] BamHI and NotI restriction sites were introduced at both ends of the primers, and TaMDH gene was used as a template for PCR amplification. Amplification conditions: 10ng DNA, 1μmol / L primers, 0.4mmol / LdNTP, 2.5U Taq DNA polymerase (Gibco Company, Grand Island, NY, USA). Denaturation at 95°C for 5 minutes, followed by 30 cycles (1 minute at 95°C, 1 minute at 55°C, 1.5 minutes at 72°C), and a final extension at 72°C for 10 minutes.
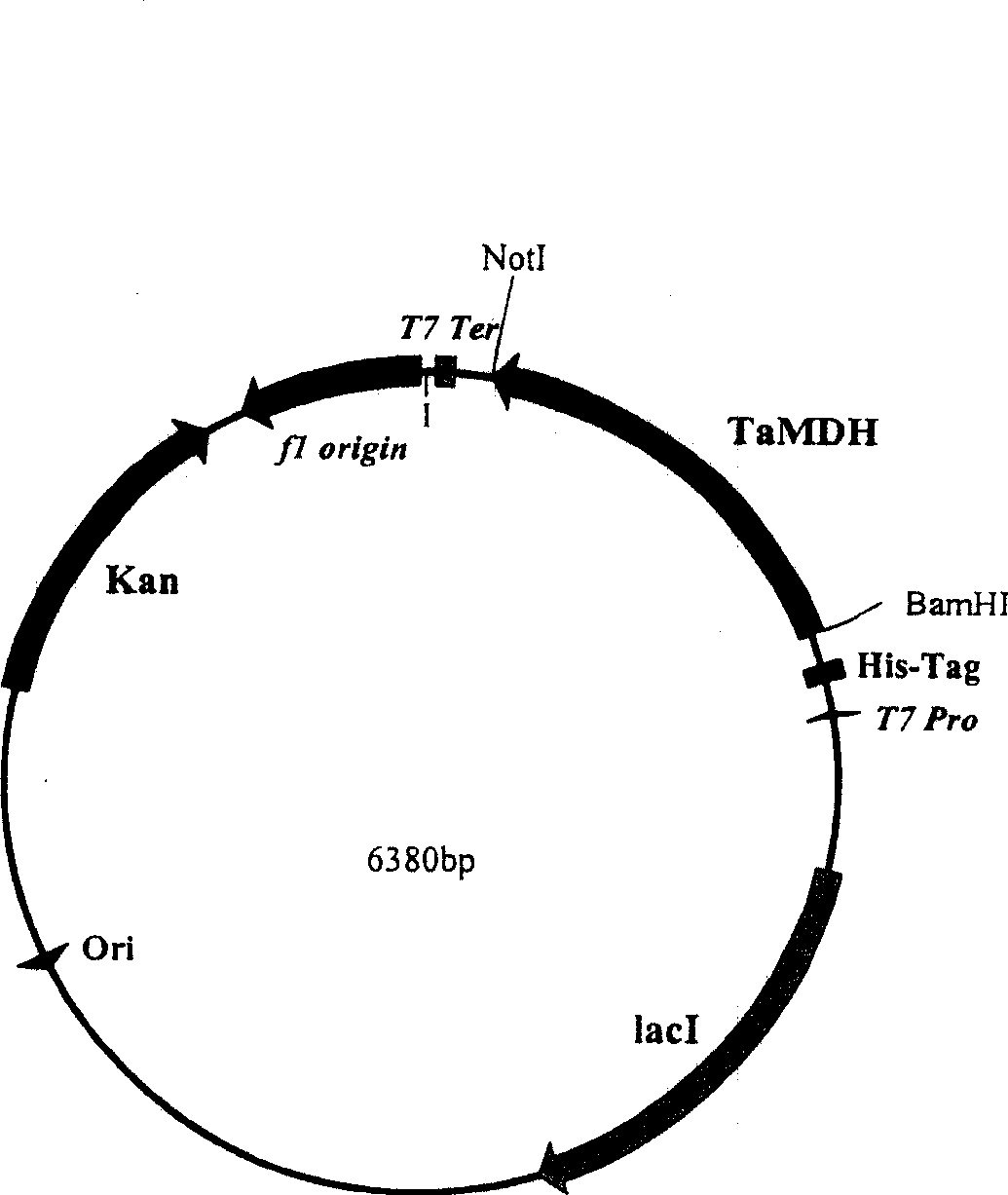
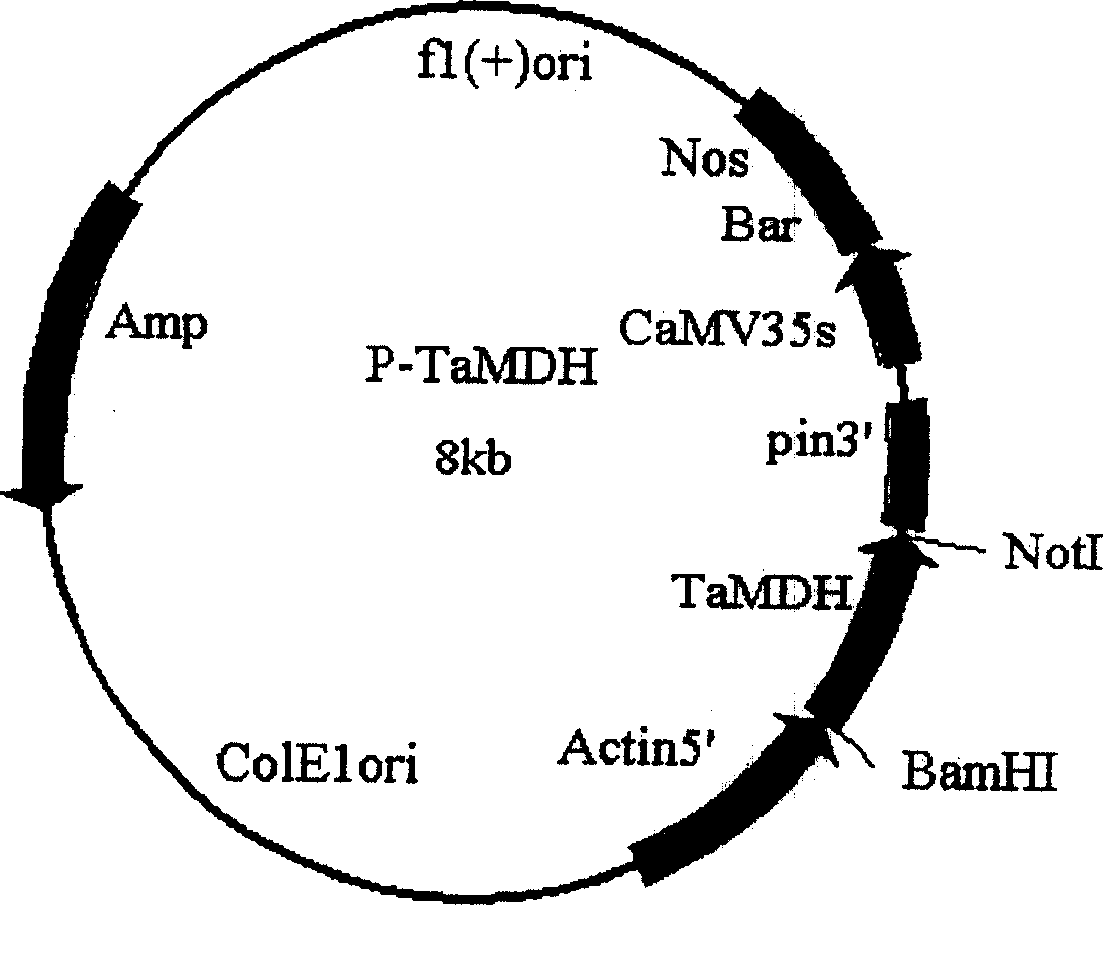
[0045] PCR products were subjected to 1.0% agarose gel electrophoresis, using DNA Isolation Kit (Gibco Company, Grand Island, NY, USA) purified the electrophoresis product, digested with BamHI and NotI double enzymes, connected to the vector pET-28a (Novagen Company, U...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com