Antisense nucleic acid of resistant and tolerant dimethoxyphenecillin staphylococcus aureus drug resistant gene
A methicillin-resistant, antisense nucleic acid technology, applied in the antisense nucleic acid field of anti-methicillin-resistant Staphylococcus aureus drug resistance gene, can solve the problem of not being a good target site, etc., to achieve the suppression of MRSA drug resistance, The effect of broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
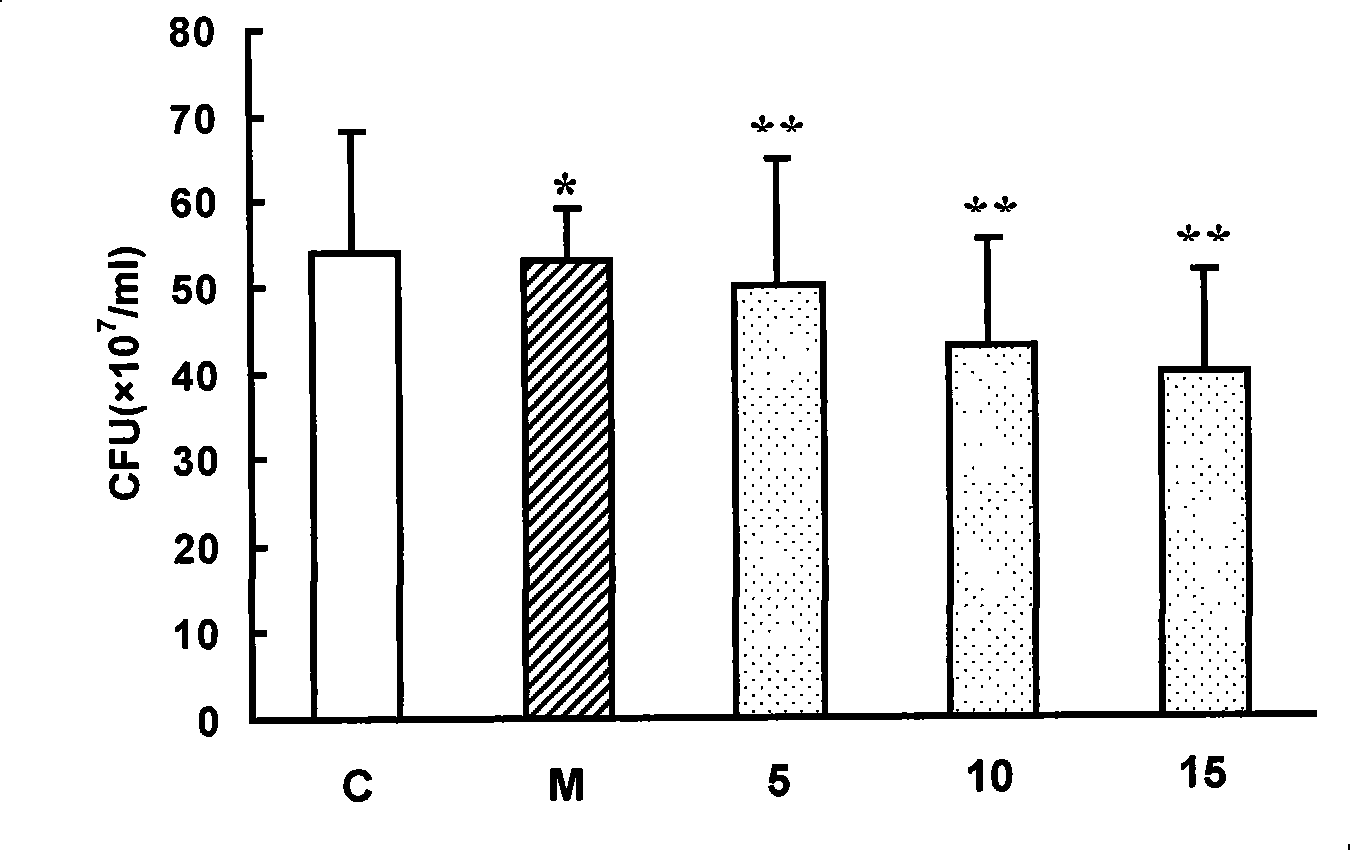
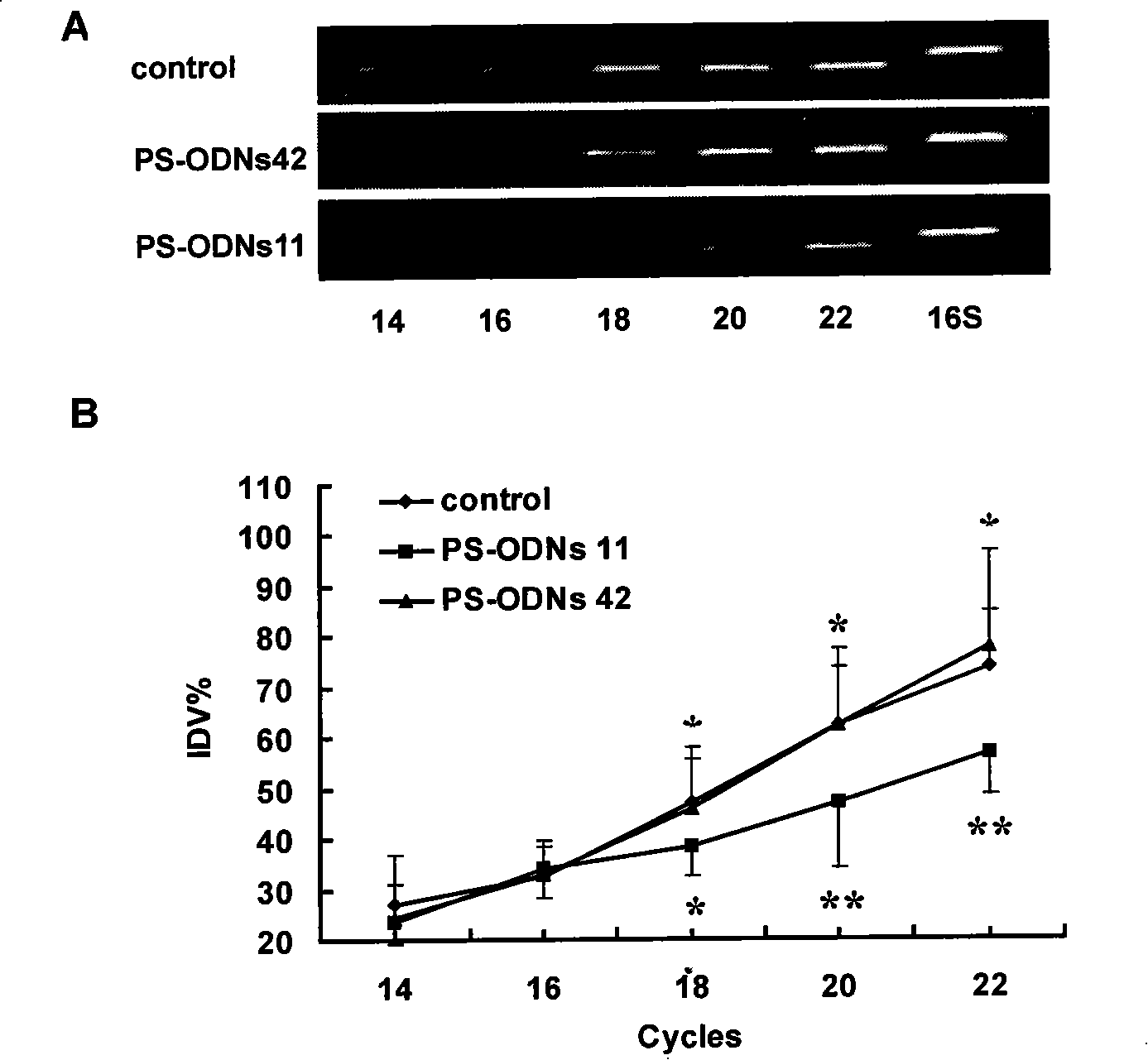
[0028] The design of antisense thio-oligodeoxynucleotides (PS-ODNs) and thio-deoxyribozyme (PS-DRz) of embodiment 1 anti-mecR1 or blaR1 mRNA:
[0029] The sequence of the drug-resistant gene mecR1 / mecA of MRSA (gene number gi:2791983) was retrieved in GENBANK. The sequence is 9047bP in full length and encodes 8 open reading frames, including 3 mec regions and 5 ORF regions. The sequence between 1615bp and 3372bp is the mecR1 gene sequence, and the sequence between 3472bp and 5478bp is the mecA gene sequence. The full length of mecR1 is 1758bp, which is the upstream gene of mecA, which can encode MecR1 protein and regulate the expression of mecA; the full length of mecA is 2007bp, which encodes the penicillin binding protein PBP2a. Using the same method, the genome sequence of blaR1 / blaZ (gene number gi: 33390917) was retrieved. The sequence is 3080 bp in full length and encodes three open reading frames, namely blaZ, blaR1 and blal. The sequence between 953bp and 2710bp is th...
Embodiment 2
[0034] Design and synthesis of embodiment 2PCR primers:
[0035] According to the gene sequences of mecR1, mecA, blaR1, blaZ and 16srRNA of MRSA reported in GeneBank, five pairs of specific amplification primers for mecR1, mecA, blaR1, blaZ and 16srRNA were designed with the software Primer Premier5.0. The primers were verified to be highly specific by BLAST software, and were synthesized by Shanghai Bioengineering Technology Service Co., Ltd. The sequences are shown in Table 2.
[0036] Table 2 designed and synthesized amplification primers
[0037]
Embodiment 3
[0038] Example 3 Preparation of electrocompetent WHO-2:
[0039] The methicillin-resistant Staphylococcus aureus strain WHO-2 was provided by the China Antimicrobial Resistance Surveillance Center. Transplant 1ml of freshly cultured overnight WHO-2 into 100ml of nutrient broth medium, culture at 37°C and 210rpm until mid-logarithmic growth, and measure its OD 600 =0.55~0.6. After 20 minutes in ice bath, subpackage (40ml / centrifuge tube), centrifuge at 6000rpm for 10min, discard the supernatant, mix and resuspend the bacterial pellet with 40ml ice-bathed deionized water, centrifuge at 6000rpm for 10min, discard the supernatant, repeat 2 times. Afterwards, the bacterial pellet was mixed and resuspended with 20 ml ice-bathed 10% glycerol, centrifuged at 6000 rpm for 10 min, and the supernatant was discarded. The bacterial pellet was mixed and resuspended with 1 ml of ice-bathed 10% glycerol, centrifuged at 6000 rpm for 10 min, the supernatant was discarded, and repeated twice....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com