Fluorescent quantitative PCR method for real-time detecting mononucleotide polymorphism and reagent kit thereof
A technique of single nucleotide polymorphism and fluorescence quantification, which is applied in the direction of measuring devices, biochemical equipment and methods, microbial determination/inspection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0048] Embodiment-hepatitis B virus (HBV) real-time detection of 1896 mutation fluorescence quantitative PCR method and its kit
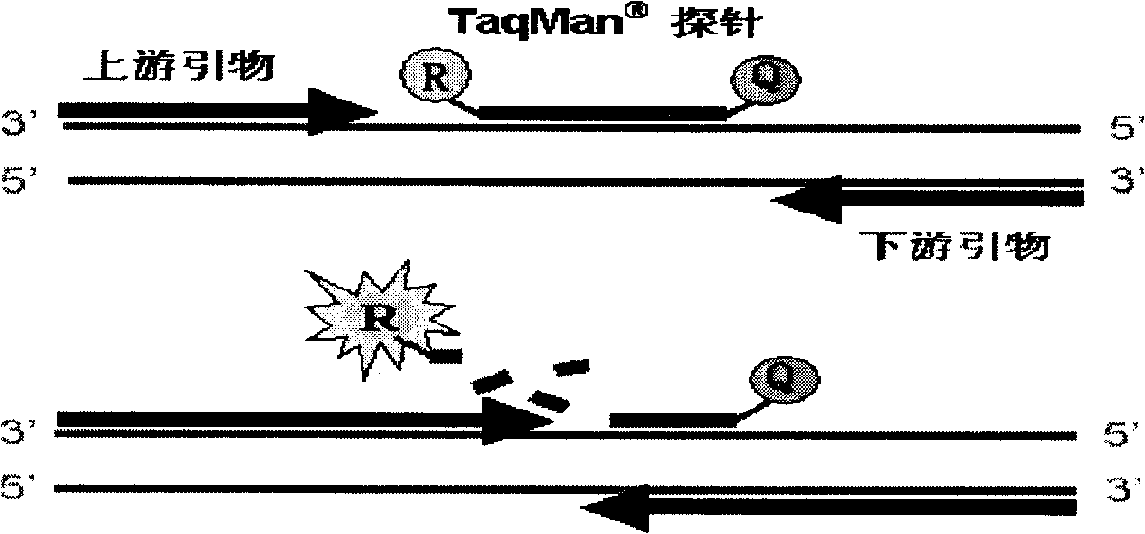
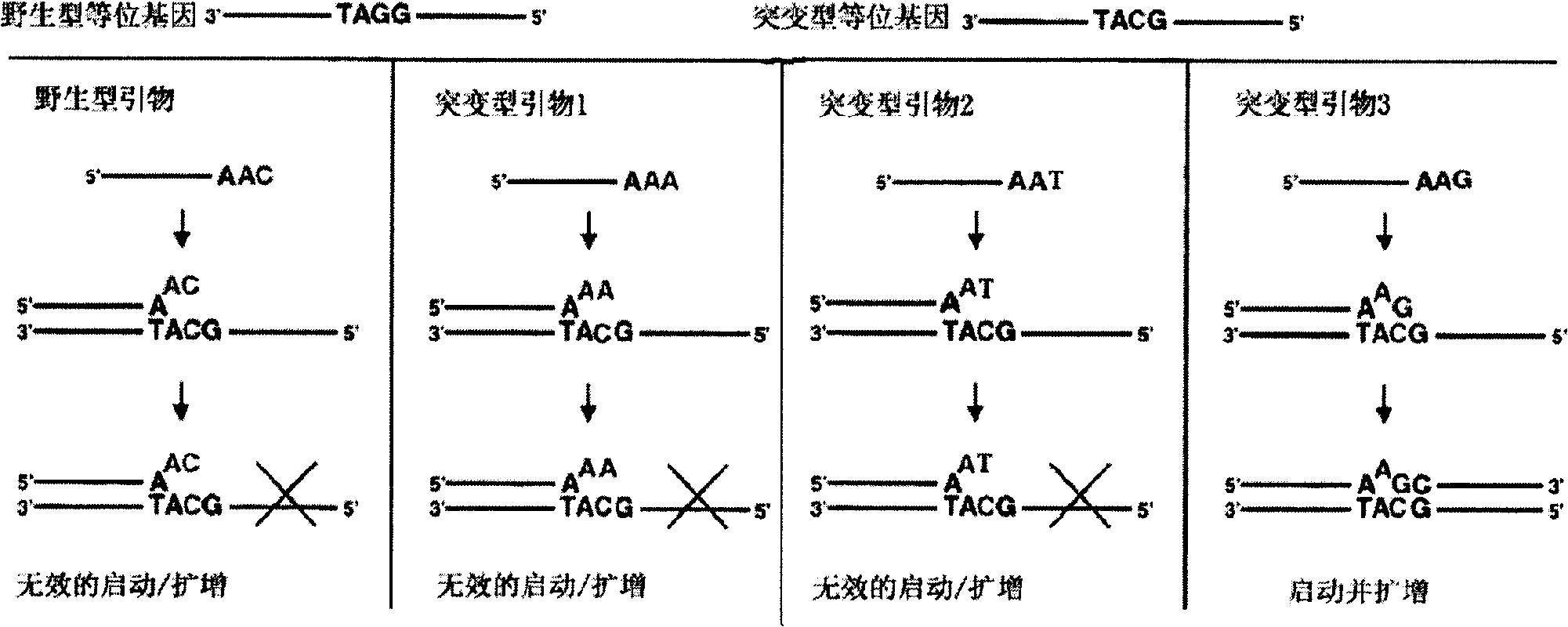
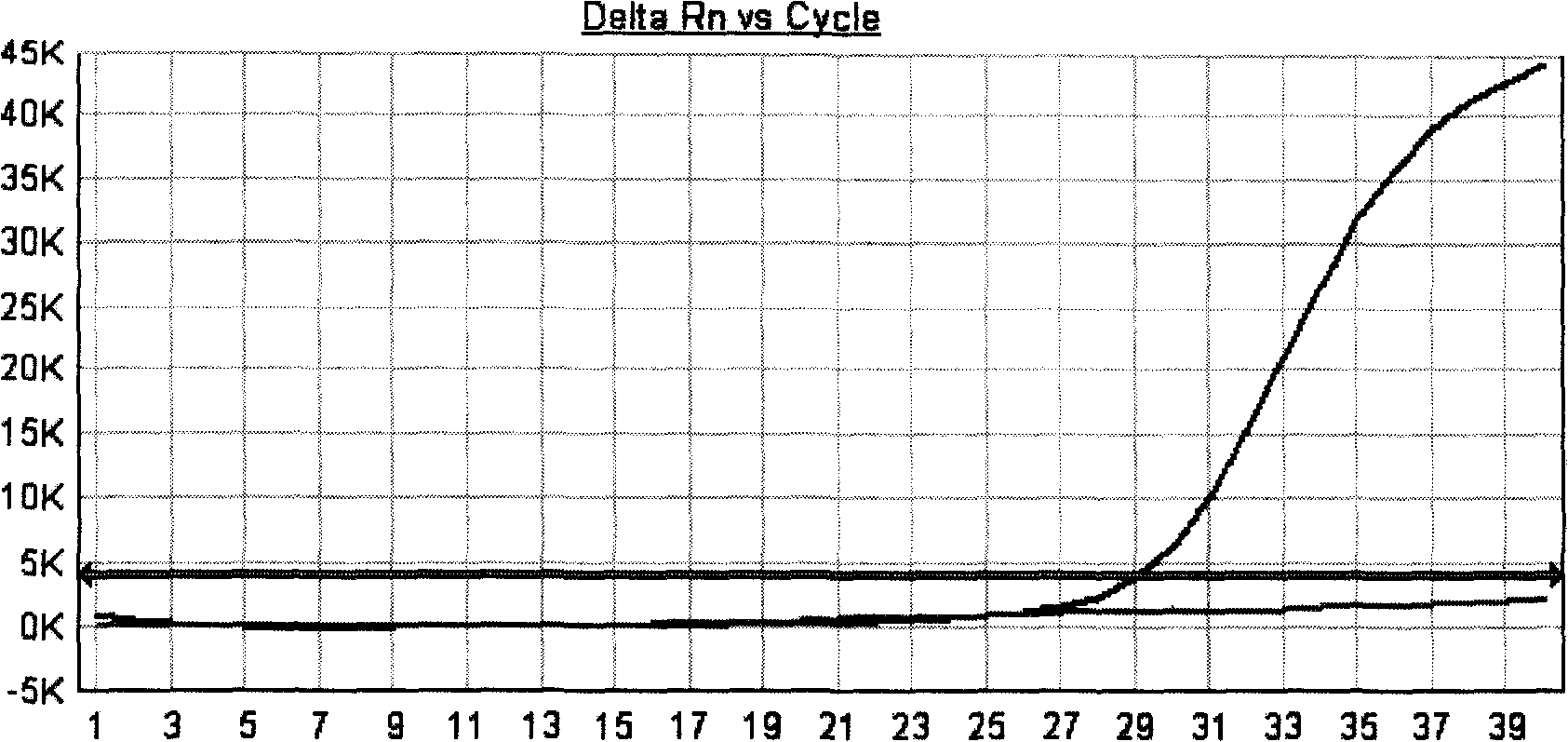
[0049] Primer design: design a sequence with a length of 16-25 bases, fix the base corresponding to the mutation site at its 3' end, the 3' end of the primer, and control the Tm value of the sequence (a complete The required temperature when half of the double-stranded DNA sequence is cleaved) meets the PCR requirements, and then the relevant primer design software is used to select another primer and probe, and two sets of positive and negative primers can be designed on the double-stranded template. Another primer and probe design-related requirements are the same as general fluorescent PCR, and the primer and probe design software are Premier primer 5.0, OLIGO 6.0, Primer Express3.0 and DNAStar, etc. The experimental results prove that the implementation of this scheme can perform SNP detection well, and at the same time, false positives in sampl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com