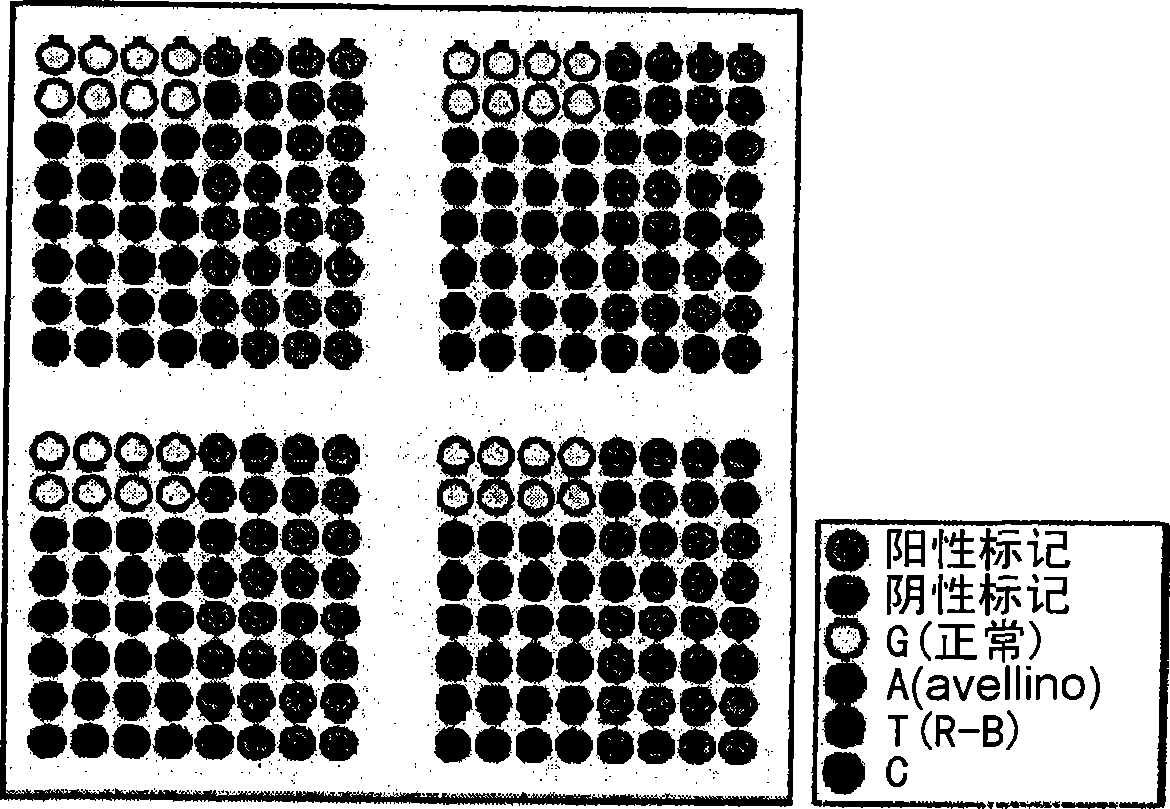
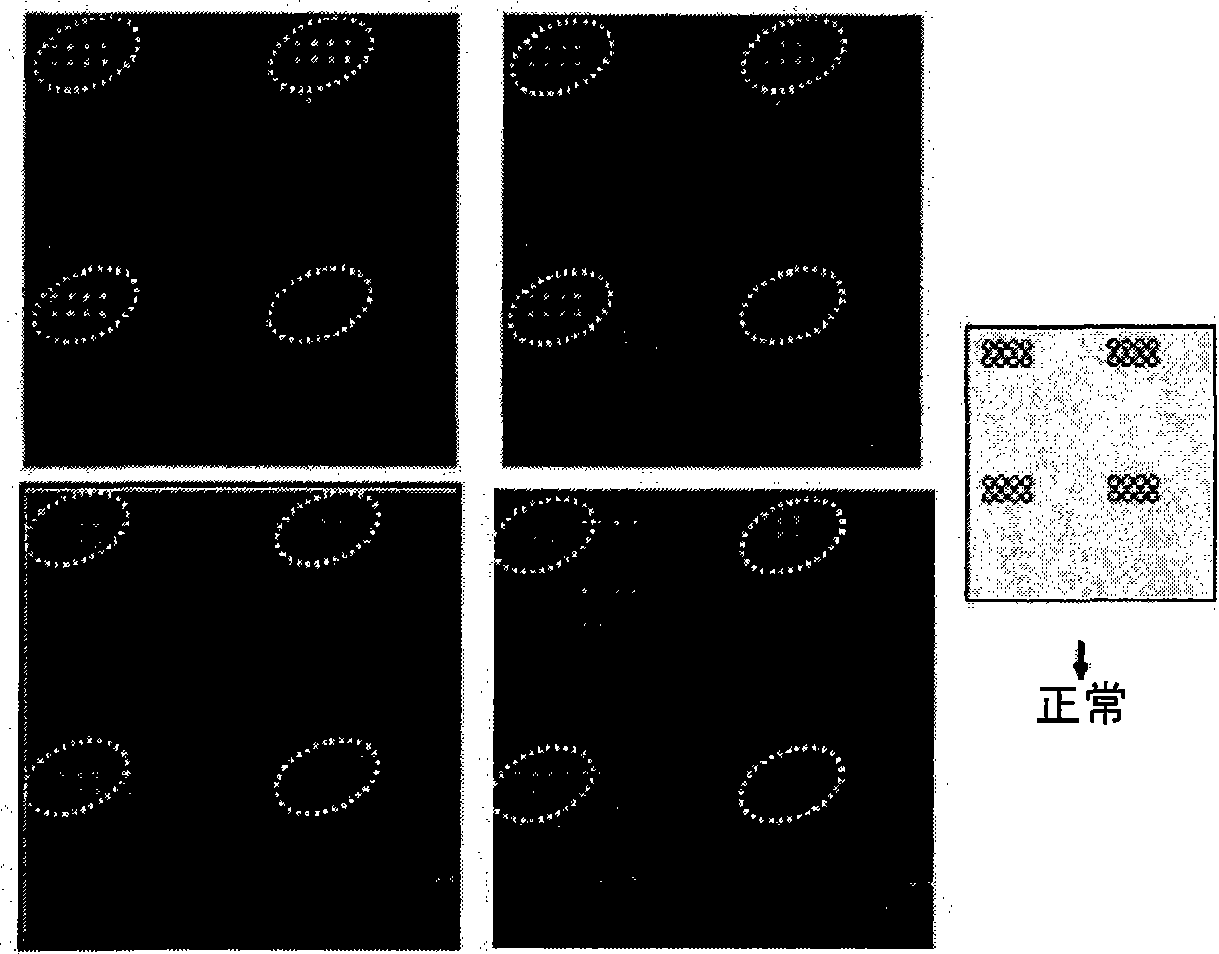
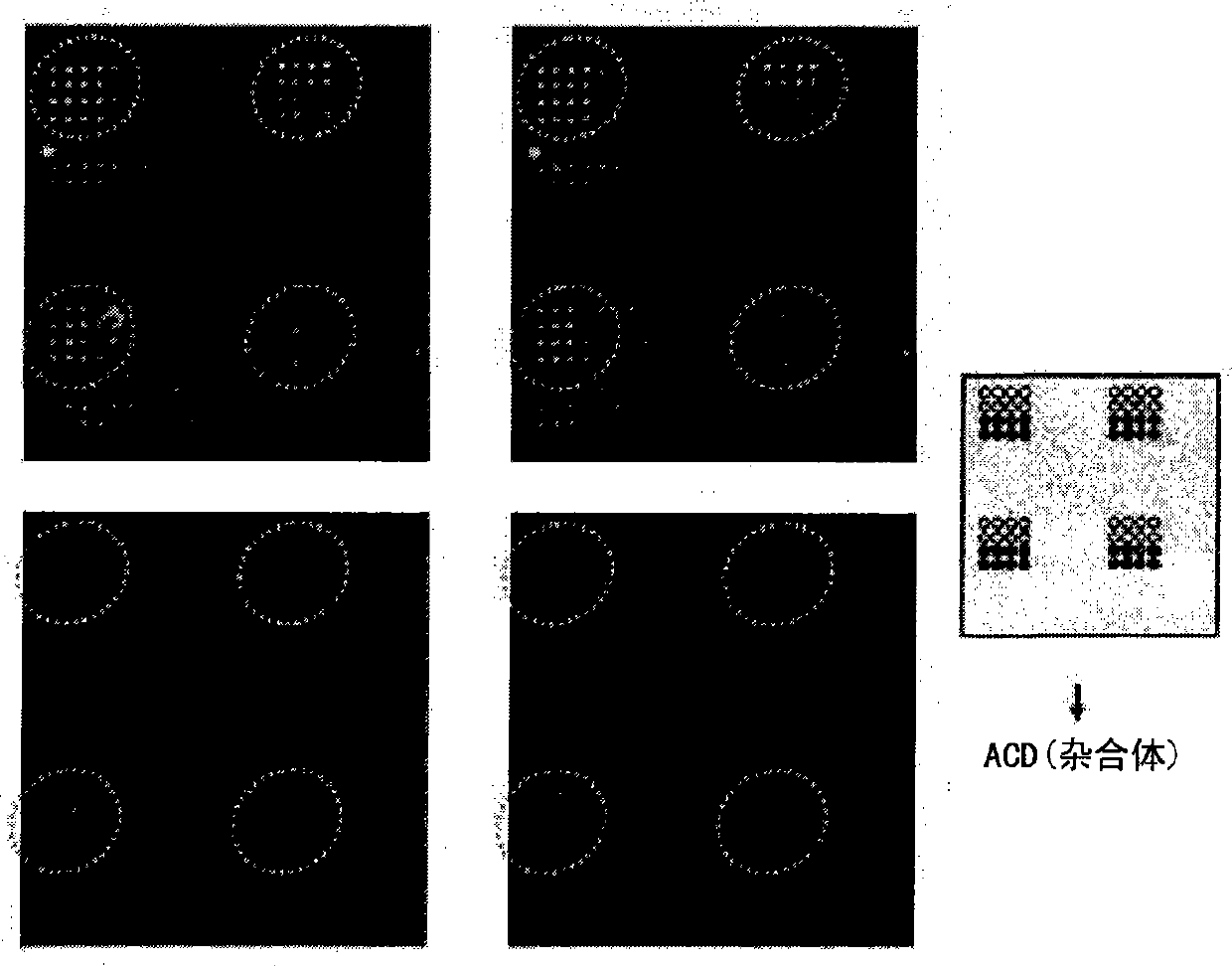
DNA chip for diagnosis of corneal dystrophy
A DNA chip, malnutrition technology, applied in the field of oligonucleotides, can solve problems such as visual loss
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0102] Example 1: Determining the mutation type of BIGH3 protein
[0103] In order to construct a diagnostic probe for determining the gene mutation of BIGH3, one of the causes of ophthalmic diseases including Avellino corneal dystrophy, its mutation site was identified to generate a probe.
[0104] The mutation sites of the BIGH3 protein were identified, and their amino acid and nucleotide sequences were guaranteed through GenBank and OMIM (Online Medelian Inheritance in Man), NCBI Nucleotide Sequence Database (National Center for Biotechnology Information), and the equivalent of each disease bit genetic information is understood. The type of mutation to be examined was determined to test the efficacy of the DNA chip. Especially among hotspots for BIGH3 gene mutations, regions of mutations causing Avellino dystrophy, Lattice I dystrophy, and Ray-Brühl I were selected (Table 1).
[0105] Table 1. Ophthalmic disorders caused by mutations in the BIGH3 gene
[0106] ...
Embodiment 2
[0108] Example 2: Obtaining the search region by PCR
[0109] To retrieve all the mutations shown in Table 1, five pairs of primers including exon 4, exon 11, exon 12 and exon 14 were designed. Among these primers, two pairs of primers were used to amplify the mutated region of exon 4. One of the two pairs of primers (primer 1 and primer 2) was considered suitable for the experiment of diagnostic DNA chip, and the other pair was designed for direct DNA sequence analysis (primer 3 and primer 4). In addition, DNA probes and their complementary primers to be searched (reverse primers) are labeled with fluorescent chemicals on their 5' hydroxyl groups. Those primer pairs were thus effectively distinguished by Cy5 and Cy3 binding to primer 2 and primer 4, respectively.
[0110] Table 2: Primers used to amplify the region of the gene including the mutation site
[0111] Primer# serial number. sequence(5'->3') 1 1 agc cct acc act ctc aa 2 2 cag gcc tc...
Embodiment 3
[0114] Example 3: Manufacturing of probes for diagnosing BIGH3 gene mutations
[0115] To retrieve the corneal dystrophy mutations shown in Table 1, probes were designed containing sequences extending 5-8 more nucleotides, preferably 7 more nucleotides, from both sides of the hotspot. Probes were designed for the normal counterparts, comprising the same sequence as above but with the nucleotide sequence mutated in the center replaced by the normal.
[0116] Table 3: Constructed probes for the diagnosis of ophthalmic disease mutations
[0117]
serial number
genotype normal /
mutation
probe sequence
Exon 11 normal R124 acg gac cgc acg gag 4 12 Avellino Malnutrition R124H acg gac cac acg gag 4 13 Ray-Buddha (CDB1) R124L acg gac ctc acg gag 4
[0118] 14 normal R124 cac gga ccg cac gga 4 15 Lattice Type I R124C cac gga ctg cac gga 4 16 normal P501 gac ccc ccc aat ggg 11 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com