Design method for realtime fluorescent quantitative PCR experiment interior label
A technology for real-time fluorescence quantification and method design, which is applied in biochemical equipment and methods, and the determination/inspection of microorganisms. The effect of a negative result
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
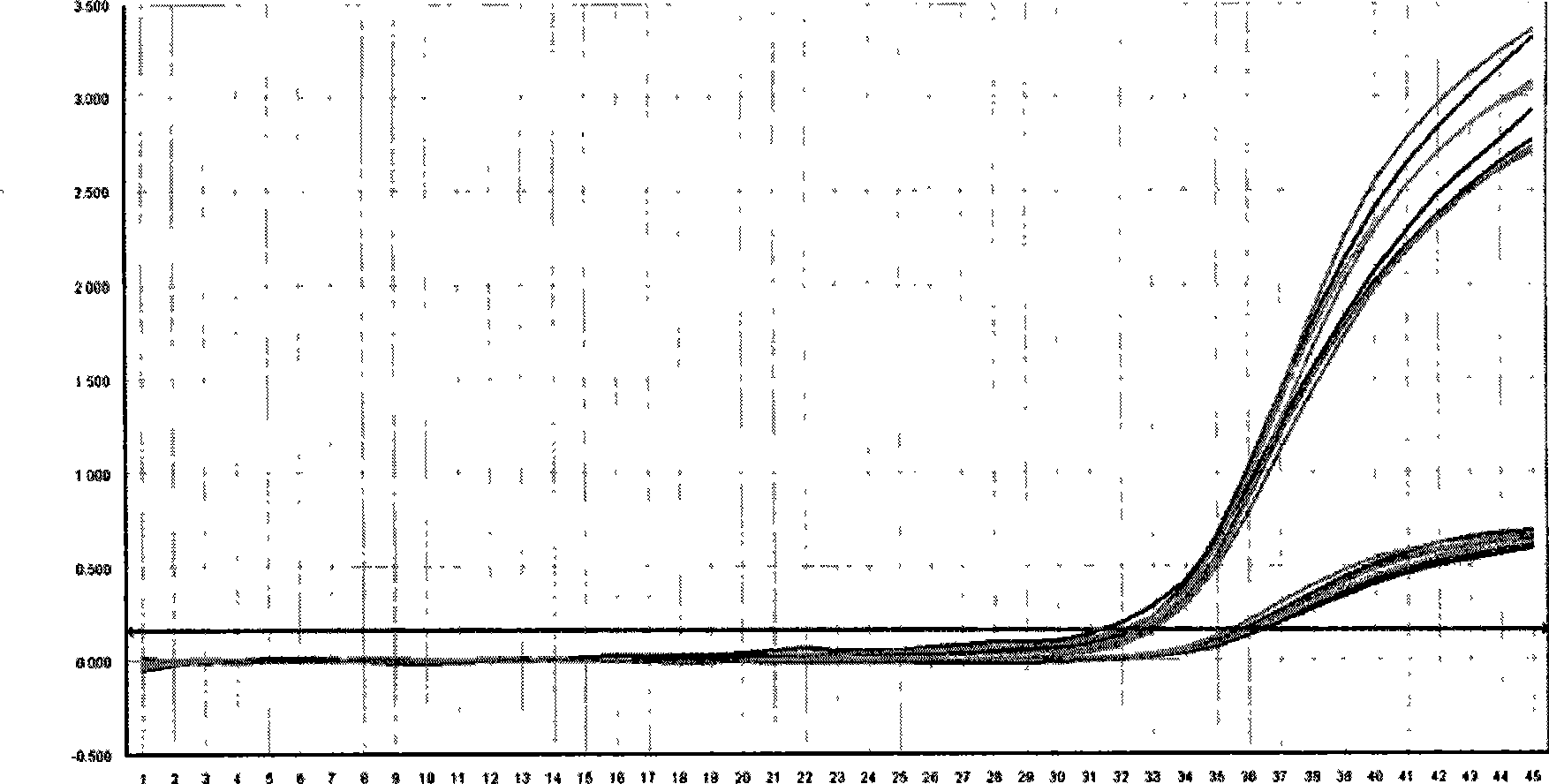
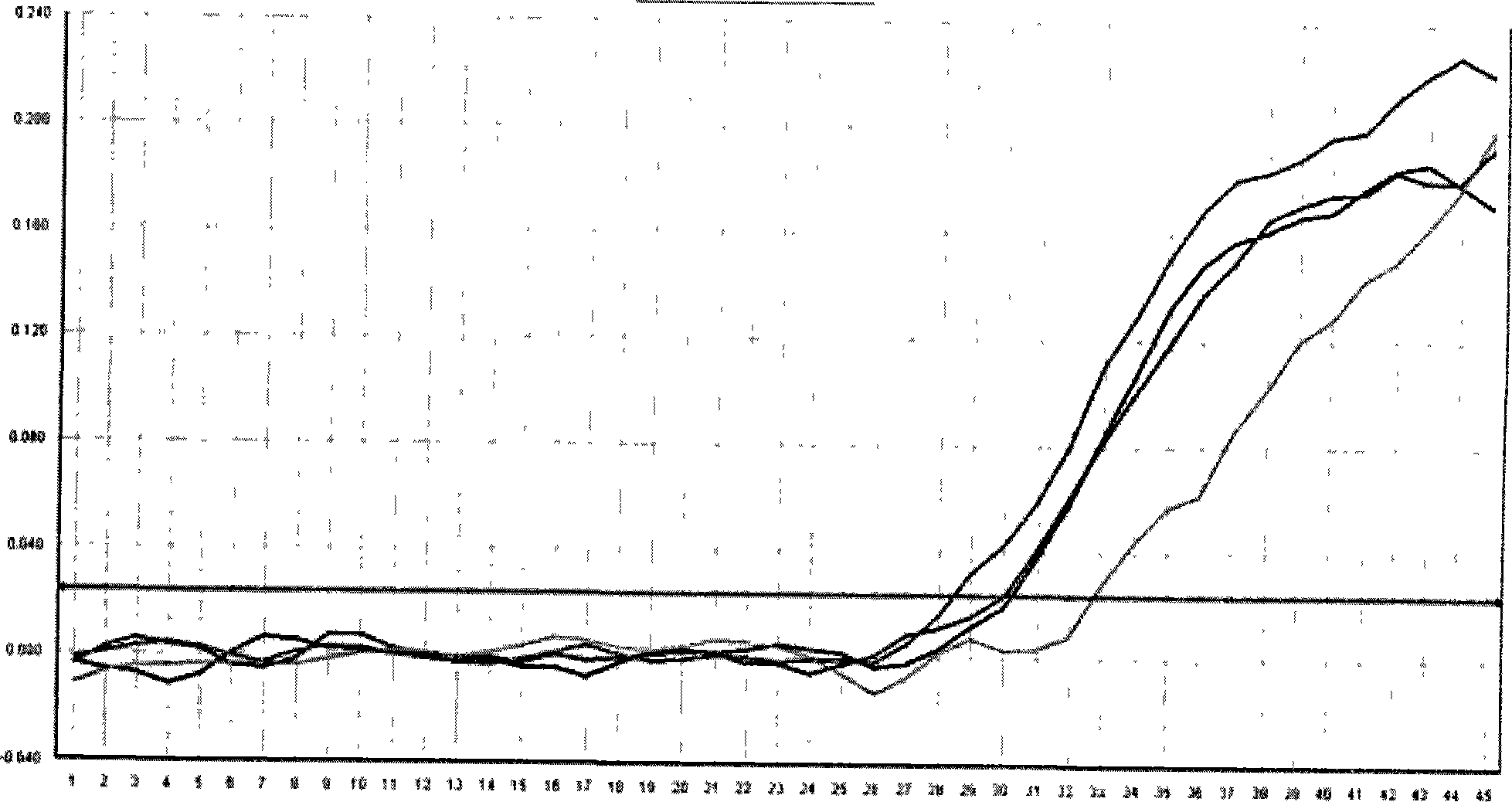
[0019] Example 1: Example of real-time fluorescence quantitative PCR detection of low-concentration hepatitis B virus deoxyribonucleic acid (HBV-DNA) in serum
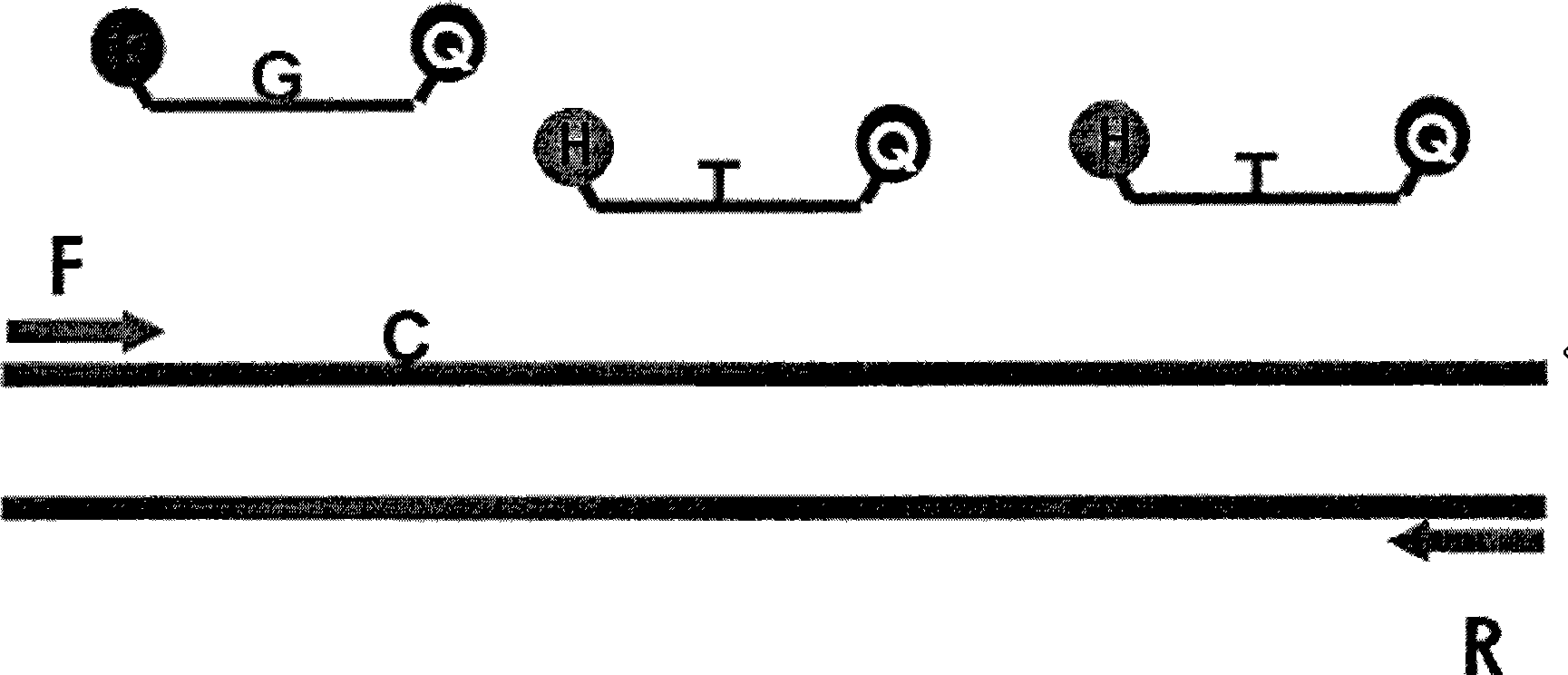
[0020] Reference figure 1 (1) Use Genbank to search for homologous segments. With the aid of primer design software, design two pairs of hepatitis B primers, namely upstream primer P1 and downstream primer P2. The sequence of upstream primer P1 is 5'-GTG TCT GCGGCG TTT TAT C-3', the downstream primer P2 sequence is 5'-ACA AAC GGG CAA CAT ACC T-3', the selected synthetic gene is an internal standard gene, and its base sequence is 5'-ACGGGAGCGGTTGGTGGTGGAAATCGTGCGTGACATTAAGA-3', which The primer sequence is the same as the target gene (same as P1, P2); (2) Design the target gene probe, the probe sequence is: 5'FAM-CAT CCTGCT GCT ATG CCT CAT CTT CTT-3', 5'end labeled FAM fluorescent report Set the probe concentration to 15pM; (3) Design two probes for the internal standard gene, the probe concentration for the internal stand...
Embodiment 2
[0022] Example 2: Example of real-time fluorescent quantitative PCR detection of high concentration of HBV-DNA in serum
[0023] The design of the target gene, internal standard, and primer probe is the same as in Example 1. The high-concentration serum sample (containing the HBV-DNA target gene, the concentration is higher than 500IU / ml, gradient dilution) and 0.3ul 10 5 IU / ml internal standard (including internal standard gene) was added to 100ul sample extract, probes for target gene and internal standard gene (concentrations of 15pM and 10pM respectively) were added to 50ul PCR reaction solution, internal standard and sample Synchronous processing, through nucleic acid extraction and purification steps to obtain high-purity nucleic acid, and perform absolute quantitative experiments on a fluorescent quantitative PCR machine (ABI7300, produced by Applied Biosystems, Inc.).
[0024] The same sample adopts the non-competitive internal standard method currently used in the market ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com