Identifying Viral Cell Tropism
一种细胞、病毒的技术,应用在HIV病毒领域,能够解决不能提供HIV-2型病毒特征测试等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
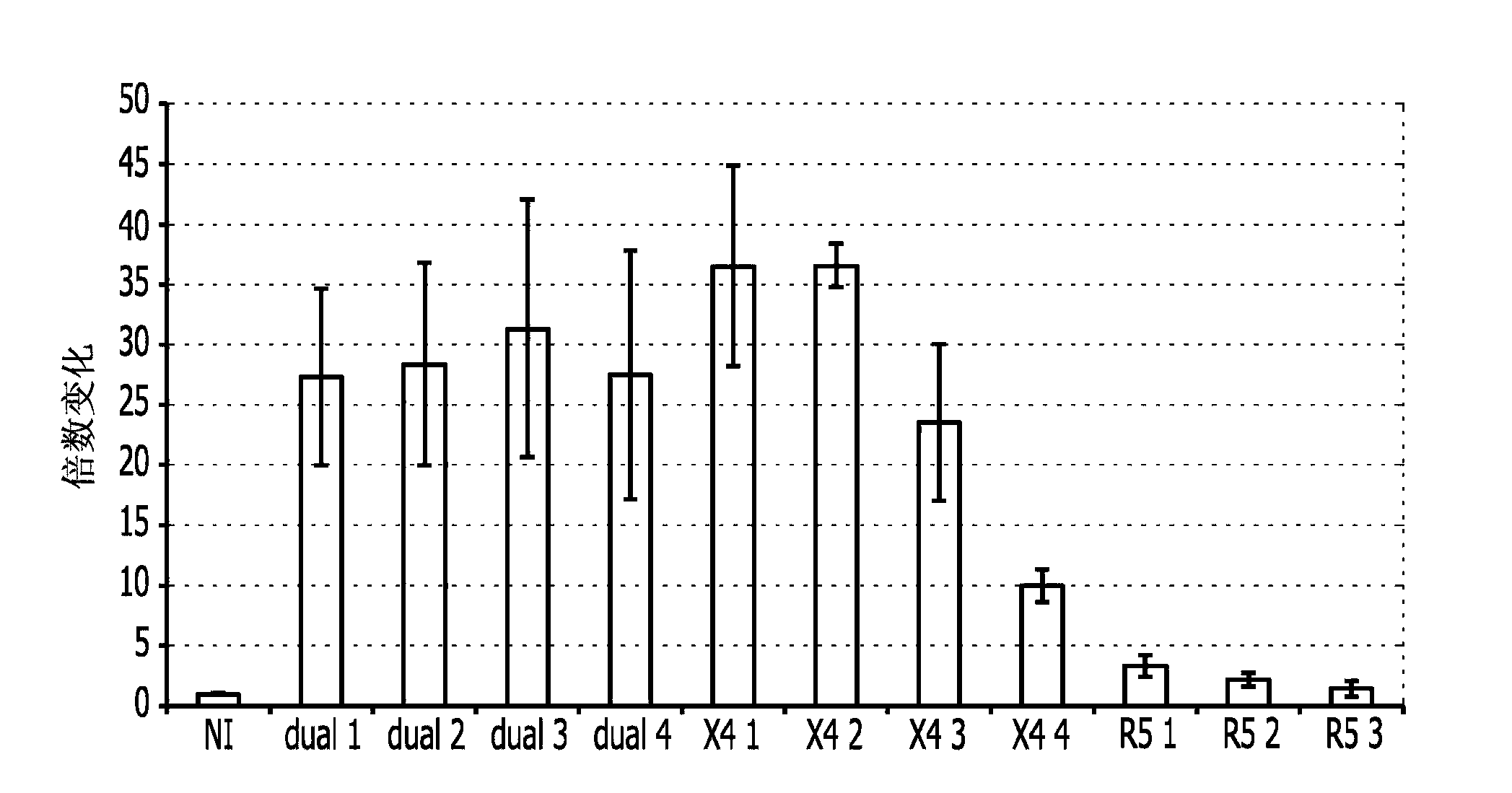
Image
Examples
Embodiment Construction
[0018] The term "miRNA" or "microRNA" refers to a class of RNAs, usually 20 to 25 nucleotides in length, involved in the transcription of certain genes by degrading or blocking the translation of mRNAs derived from those transcriptions post adjustment. A "target mRNA" of a miRNA refers to an mRNA known or determined to be degraded or translationally blocked by said miRNA. miRNA in Griffiths-Jones ((2004) Nucleic Acids Res.32:D109-D111), Griffiths-Jones et al. ((2008) Nucleic Acids Res 36:D154-D158) and miRA database (miRBase, http: / / microRNA. specifically described in sanger.ac.uk).
[0019] The expression "virus" as used herein includes all types of viruses. In particular, the virus may be selected from the group of more or less pathogenic viruses of their variants or varieties, such as retroviruses, especially HIV viruses, influenza viruses, coronaviruses, measles viruses, herpes viruses (including EBV , herpes simplex virus and CMV virus), papillomavirus. Preferably, th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com