Establishment of female genital cancer relevant SNP sites multiple detection method
A cancer-related, genital technology, applied in the field of biotechnology applications, can solve problems such as difficulty in further increasing the number of sites and limitations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach 1
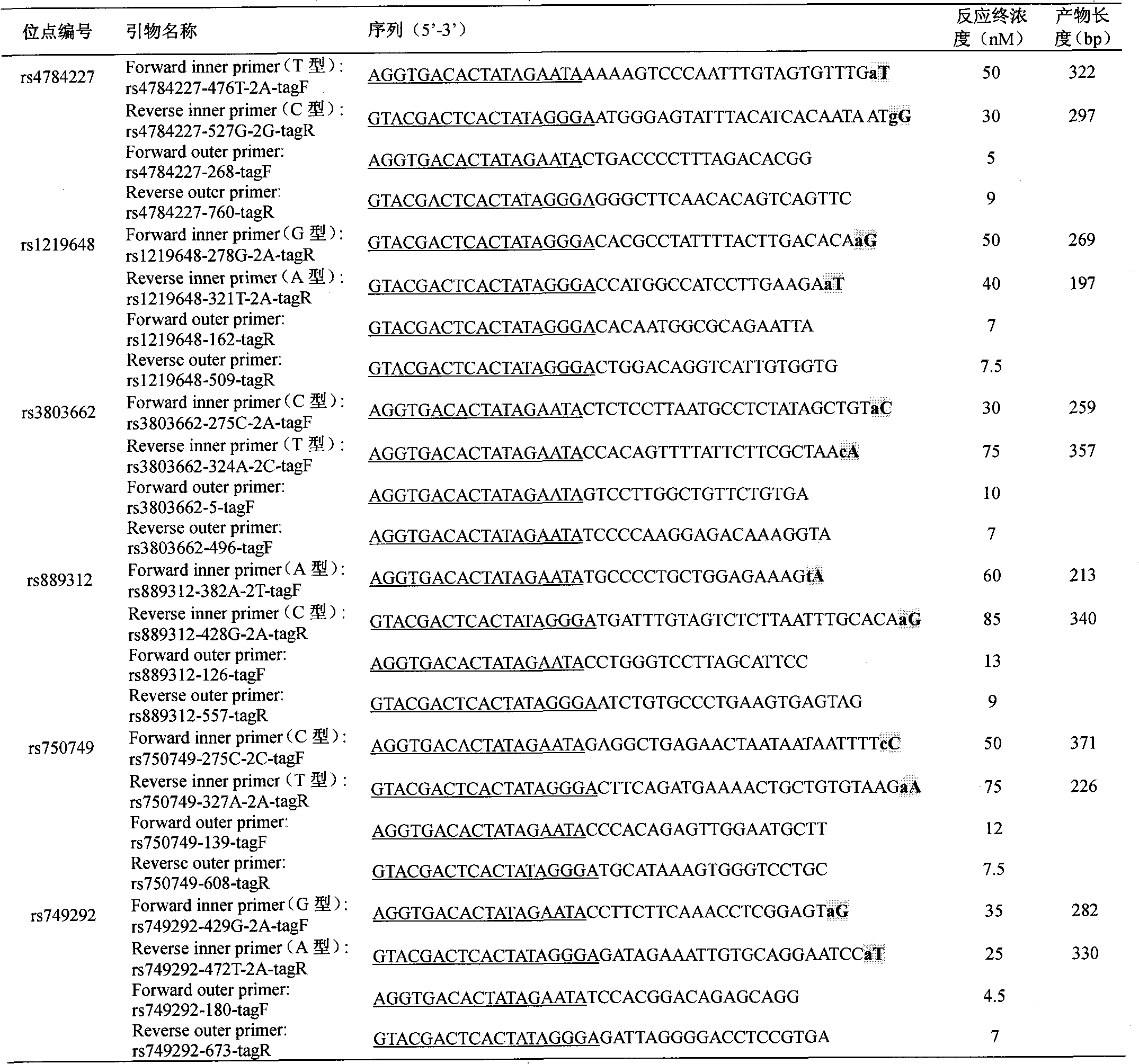
[0014] Embodiment 1: multiplex PCR amplification
[0015] A Multiplex PCR Kit (QIAGEN) kit was used. The PCR reaction system was 20 μl, including 0.5 μl of whole blood DNA template, 2 μl of buccal swab DNA template, 500 nM of universal primers, and the concentrations of other primers are shown in Table 1. The PCR reaction is divided into 6 steps to amplify the target fragment: Step 1: Pre-denaturation at 95°C for 10 minutes; Step 2: Denaturation at 95°C for 30 seconds, renaturation at 60°C for 30 seconds, extension at 72°C for 45 seconds, cycle 3 times; Step 3: 95°C Denaturation for 30s, 58°C for 30s, 72°C for 45s, cycle 10 times; Step 4: 95°C for 30s, 68°C for 30s, 72°C for 45s, cycle 20 times; Step 5: 95°C for 30s , renatured at 55°C for 30s, extended at 72°C for 45s, and cycled 12 times; step 6: extended at 72°C for 5min, and stored at 4°C. Two replicates were made for each specimen.
Embodiment approach 2
[0016] Embodiment 2: Capillary Electrophoresis Separation
[0017] use system (Qiagen) capillary electrophoresis system, DNA high-resolution cartridge (Qiagen) and OH700 method were used as separation conditions, used in conjunction with QX Alignment Marker 15bp / 500bp, QX DNA Size Marker 25-450bp, and analyzed by Biocalculator software (Qiagen) to determine whether there were products of corresponding length. SNP typing.
Embodiment approach 3
[0018] Embodiment 3: Sequencing Verification
[0019] PCR reaction system 25 μl, including 0.5 μl whole blood DNA template, 2 μl buccal swab DNA template, Premix Ex Version 2.0 (TakKaRa) 12.5μl, take 500nM of outer primer without Tag for each site. The PCR reaction was: pre-denaturation at 95°C for 5 minutes; denaturation at 95°C for 30 s, annealing at 58°C for 30 s, extension at 72°C for 45 s, and 45 cycles; extension at 72°C for 5 min, and storage at 4°C. The products were sequenced, and the sequencing results were compared with the results of M-T-ARMS-PCR electrophoresis.
[0020]
[0021]
[0022]
[0023]
[0024]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com