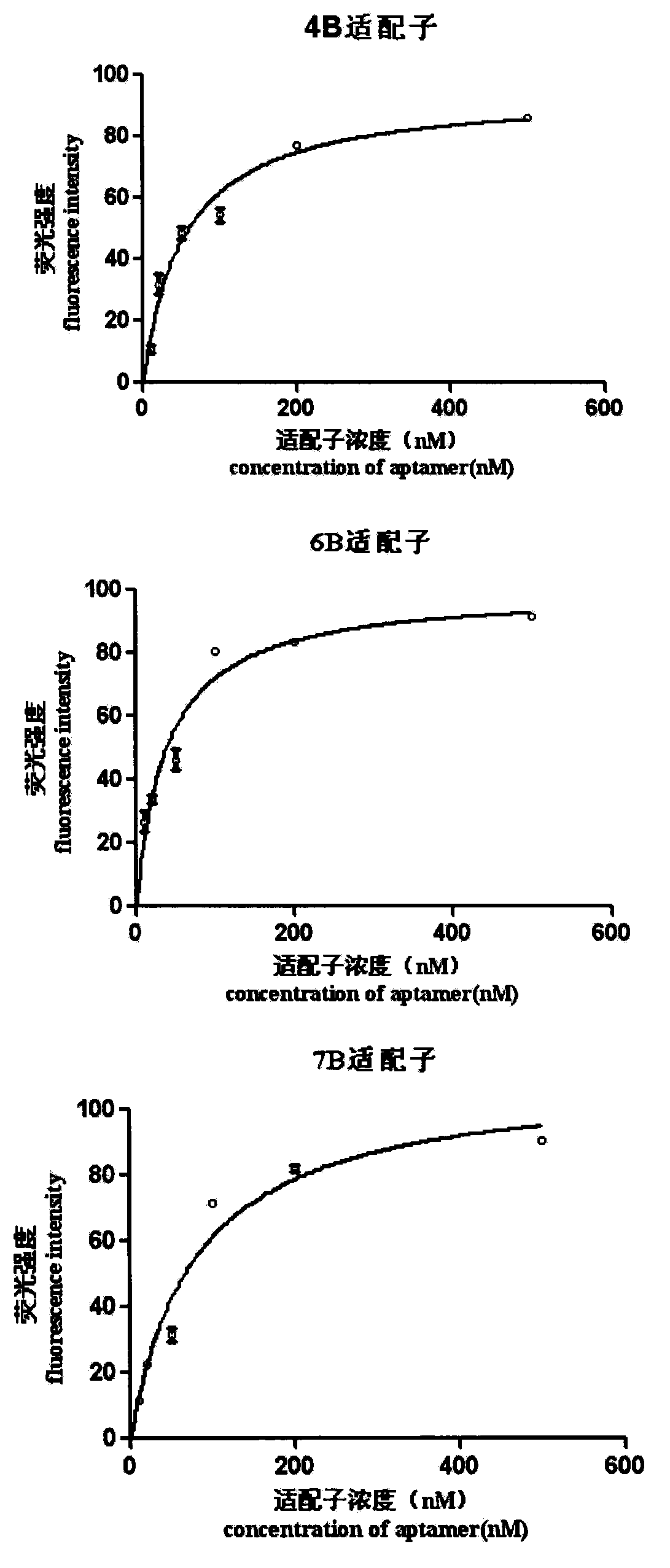
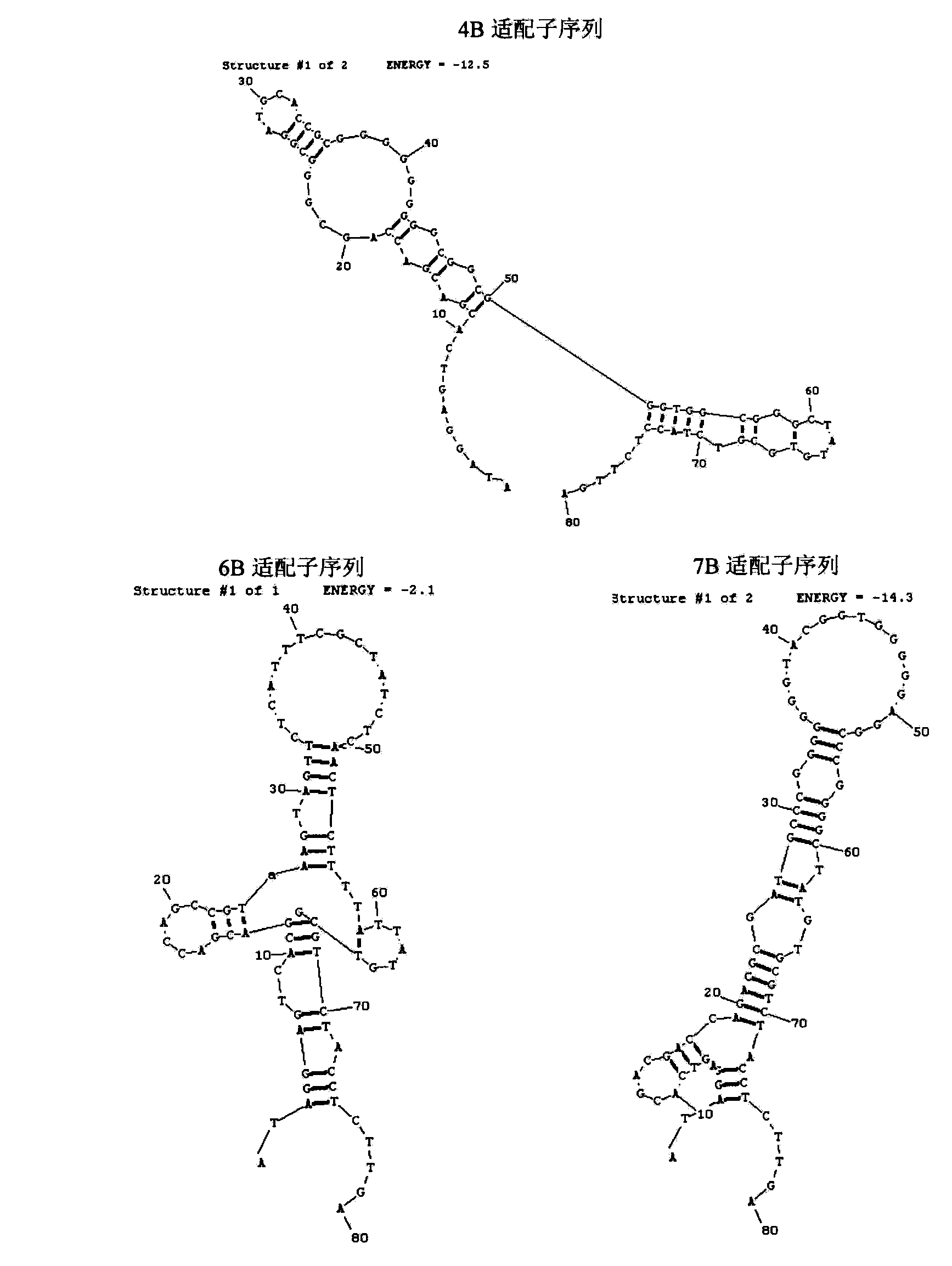
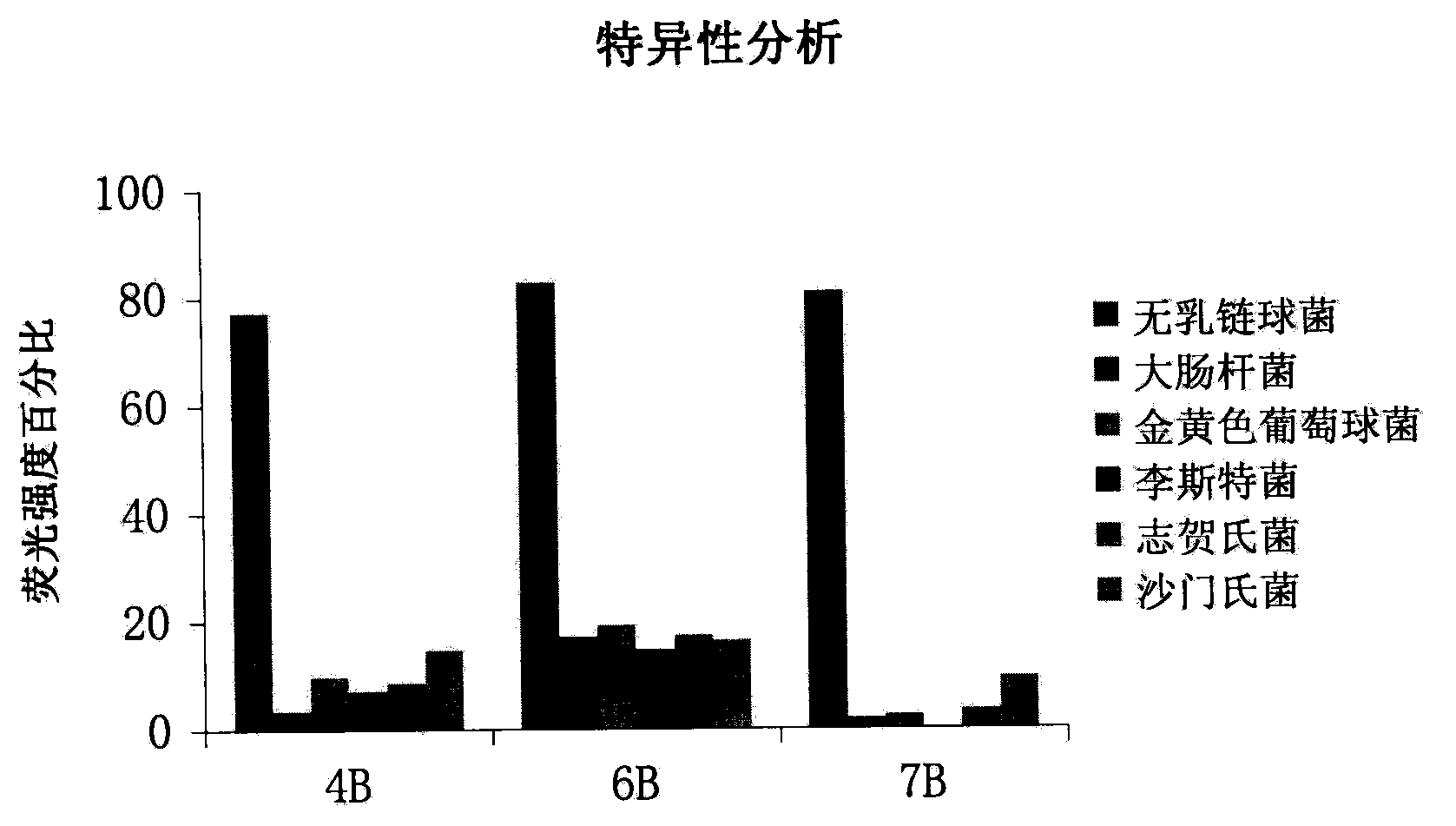
A set of oligonucleotide aptamers capable of specifically recognizing streptococcus agalactiae
A technology of Streptococcus lactis and oligonucleotides, applied in the biological field, can solve time-consuming and arduous problems, and achieve the effect of low preparation cost, short cycle and stable single-stranded oligonucleotides
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0016] Example 1: Competition SELEX screening of Streptococcus agalactiae-specific binding oligonucleotide aptamers
[0017] 1. In vitro chemical synthesis of initial random single-stranded DNA (ssDNA) library and primers (completed by Integrated DNA Technologies, USA), the sequence is as follows: 5'-ATAGGAGTCACGACGACCAG(40N)TATGTGCGTCTACCTCTTGA-3'; a random ssDNA library with a length of 80nt was constructed, The two ends are fixed primer sequences, the middle is a random sequence of 40 bases, and the library capacity is 10 14 above;
[0018] Primer I: 5'-ATAGGAGTCACGACGACCAG-3';
[0019] Primer II: 5'-TCAAGAGGTAGACGCACATA-3';
[0020] 5'phosphorylation downstream primer: 5'-P-TCAAGAGGTAGACGCACATA-3';
[0021] The random ssDNA library and the two primers were made into a 100 μM stock solution in TE buffer and stored at -20°C for future use.
[0022] 2. Conditions for PCR amplification and Lambda exonuclease digestion to prepare single-strand sub-libraries
[0023] The sy...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com