A kind of nested PCR detection method of Ralstonia solanacearum in peanut
A R. solanacearum and detection method technology, which is applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problem of high sensitivity, and achieve the effects of high sensitivity, good practicability, and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1 Nested PCR Detection of Ralstonia solanacearum in Peanut
[0030] 1. Sample DNA extraction
[0031] 1.1 Extraction of cell DNA
[0032] Inoculate peanut solanacearum in NA liquid medium, culture overnight at 28°C and 200rpm, and extract bacterial genomic DNA by CTAB method. The specific operation is as follows:
[0033] 1) Take 1.5ml culture in a 2.0ml centrifuge tube and centrifuge at 12000rpm for 2min;
[0034] 2) Discard the supernatant, add 567μl TE buffer to resuspend the cells, add 30μl 10% SDS solution and 3μl 20mg / ml proteinase K, mix gently, and incubate at 37℃ for 1h;
[0035] 3) Add 100 μl 5mol / L NaCl solution, mix thoroughly, then add 80 μl CTAB / NaCl solution, mix and incubate at 65°C for 10 minutes;
[0036] 4) Add 800μl of phenol / chloroform / isoamyl alcohol solution (the volume ratio of the three is 25:24:1), mix well and centrifuge at 12000rpm for 5min;
[0037] 5) Transfer the supernatant to a new 1.5ml centrifuge tube, add 0.6-0.8 tim...
Embodiment 2
[0063] Example 2 Conventional PCR Sensitivity Test of Peanut Solanacearum
[0064] Genomic DNA of R. solanacearum was diluted to 8 concentration gradients of 10ng / μl, 1ng / μl, 100pg / μl, 10pg / μl, 1pg / μl, 100fg / μl, 10 fg / μl and 1 fg / μl respectively. Take 1 μl of DNA of each concentration as a template, and only use specific primers W1 / W2 to carry out PCR amplification once respectively, and the same results are obtained after 3 repetitions.
[0065] figure 2 It is the amplification result of conventional PCR sensitivity detection of R. solanacearum. Among them, M is the 2000 bp DNA molecular weight marker, 1-8 are different concentrations of R. solanacearum DNA, the concentrations are 10 ng, 1 ng, 100 pg, 10 pg, 1 pg, 100 fg, 10 fg, 1 fg / μl , 9 is the negative control.
[0066] figure 2 The results showed that a single PCR amplification using specific primers W1 / W2 alone amplified a 374bp specific band in lanes 1-5, indicating that the minimum detection concentration of p...
Embodiment 3
[0067] Example 3 Nested PCR Sensitivity Test of Ralstonia solanacearum in Peanut
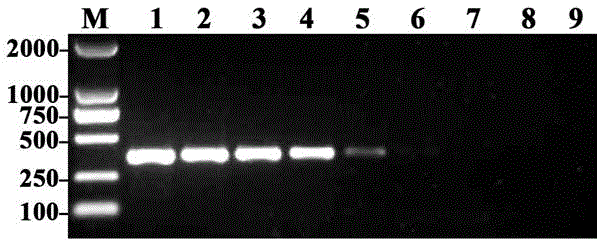
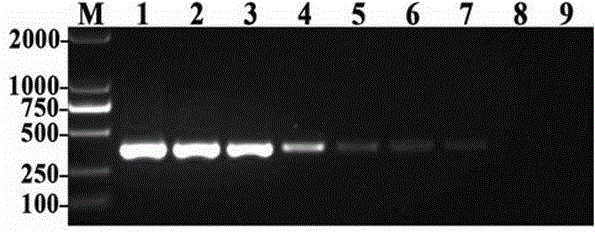
[0068] As in Example 2, the genomic DNA of Ralstonia solanacearum was diluted to 8 samples of 10ng / μl, 1ng / μl, 100pg / μl, 10pg / μl, 1pg / μl, 100fg / μl, 10fg / μl and 1fg / μl respectively. Concentration gradient. Take 1 μl of DNA at each concentration as a template, and perform the first and second rounds of PCR amplification sequentially according to Step 2 and Step 3 of Example 1, respectively.
[0069] The electrophoresis results of the products amplified by two rounds of PCR are shown in image 3 . In the figure, M is a 2000bp DNA molecular weight marker, 1-8 are different concentrations of R. solanacearum DNA, the concentrations are 10ng, 1ng, 100 pg, 10 pg, 1pg, 100 fg, 10 fg, 1 fg / μl, 9 is a negative control.
[0070] image 3 The results showed that after two rounds of PCR amplification, a specific band of 374bp was amplified in lanes 1-7, indicating that the minimum detection concentrati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com