Three microRNAs derived from tea tree and their applications
A technology for tea tree and diabetes, applied in the field of microRNA and its application, can solve the problem of detection of extremely low abundance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
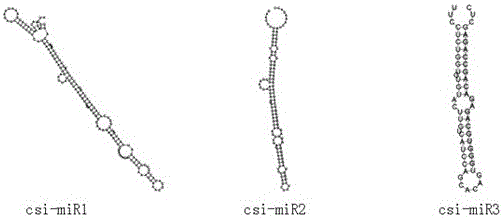
[0021] Example 1: Screening and identification of miRNA
[0022] Fresh tea tree leaves were selected for feeding mice and miRNA sequencing (2 biological replicates). Two groups of ICR mice (18-20 g), 24 in total, were fasted for 12 hours respectively, the control group was fed mouse feed, and the experimental group was force-fed tea tree leaves. Liver tissue and serum of corresponding mice were collected at different time points, and total RNA was extracted respectively. Mix equal amounts of serum sample RNA at 0h and 3h in the mouse feed group for one miRNA sequencing, and mix equal amounts of serum sample RNA at 3h and 6h for one miRNA sequencing; One miRNA sequencing was performed by mixing, and equal amounts of liver sample RNAs from 3h and 6h were mixed for one miRNA sequencing. The sample collection and sequencing of the force-fed tea tree leaf group were as above. According to the sequencing results, it was selected to be found in the tea tree leaf miRNA sequencing a...
Embodiment 2
[0024] Example 2: Distribution of 3 miRNAs in plants
[0025] The improved CTAB method was used to extract and enrich small RNAs from tea tree leaves. The details are as follows: (1) Take 0.1 g of young leaves of tea tree from each sample, grind them quickly in liquid nitrogen, and transfer them to 600 µl CTAB buffer (2% CTAB; 100 mmol / L Tris-HCl, pH 8.0; 20 mmol / L EDTA; 1.4 mol / L NaCl; 2% β-mercaptoethanol) in a 1.5 ml centrifuge tube, mix well, and place in a 65°C water bath for 20 min. (2) Centrifuge at 8 000 r / min at 4°C for 10 min, transfer the supernatant to a new centrifuge tube, and extract twice with chloroform / isoamyl alcohol (24:1). (3) The supernatant was transferred to a new centrifuge tube again, 1 / 4 volume of 10 mol / L LiCl was added, mixed well, and precipitated at -20°C for 2 h. (4) Centrifuge at 10 000 r / min at 4°C for 10 min, discard the supernatant, dissolve the precipitate with 300 µl DEPC water, add 50 µl 50% PEG8000 and 50 µl 5 mol / L NaCl, mix well, and...
Embodiment 3
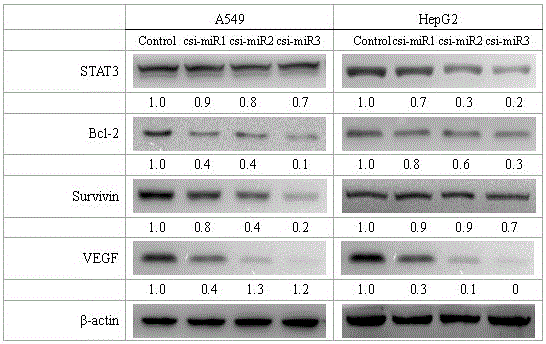
[0031] Embodiment 3: antitumor efficacy test
[0032] 1. In Vitro Antitumor Experiment
[0033] 1.1 The impact on the value-added of HepG2
[0034] Take a bottle of cultured cells in the exponential growth phase for 3 to 4 days, add an appropriate amount of 0.25% Trypsin-EDTA solution to make the adherent cells fall off, and use RPMI 1640 medium with 10% embryonic bovine serum to make 1×10 4 cells / ml suspension. Take a 96-well plate, add 100 μL of cell suspension to each well, and place at 37°C, 5% CO 2 and 100% humidity incubator for 24 hours, each well was added with different concentrations of csi-miR1, csi-miR2, and csi-miR3 synthesized by chemical methods, and each concentration was set to 5 duplicate wells, and solvent control group and positive Control group (paclitaxel). Continue to culture in the incubator for 48 h. For MTT, use serum-free RPMI1640 medium to make a 1 mg / ml solution, add 50 μl to each well, incubate at 37°C for 4 hours, remove the supernatant, add...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com