Long rigid spacers to enhance binding kinetics in immunoassays
A technology of spacers and binding molecules, applied in the field of devices for target molecules
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0265] Example 1 – Using long dsDNA as an adapter
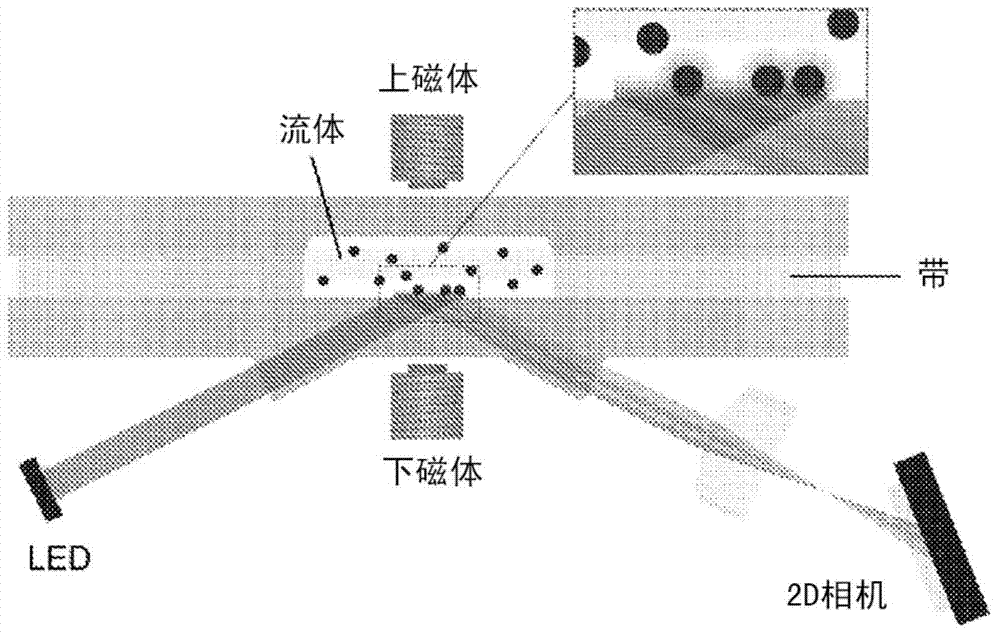
[0266] The length (extension) of the linker was analyzed in one set of experiments. For this reason, it is a very rigid molecule and thus also leads to long dsDNA extending far from the surface to serve as linkers. Specifically, 500 nm streptavidin-coated particles were incubated with dsDNA of different lengths. Each DNA molecule contains a biotin moiety at one end of the DNA molecule and a Texas Red molecule at the other end of the molecule. A solution containing particles and DNA was injected into the cartridge and bound to the sensor surface, coated with anti-Texas Red antibody using magnetic attraction. After a magnetic washing step that removes unbound particles from the surface, the number of particles bound to the surface is determined.
[0267] The binding kinetics of the particles to the surface was found to be significantly enhanced. For example, for magnetic nanoparticles with an average diameter of ~500 nm, ...
Embodiment 2
[0269] Example 2 – Effect of linker surface density on binding kinetics
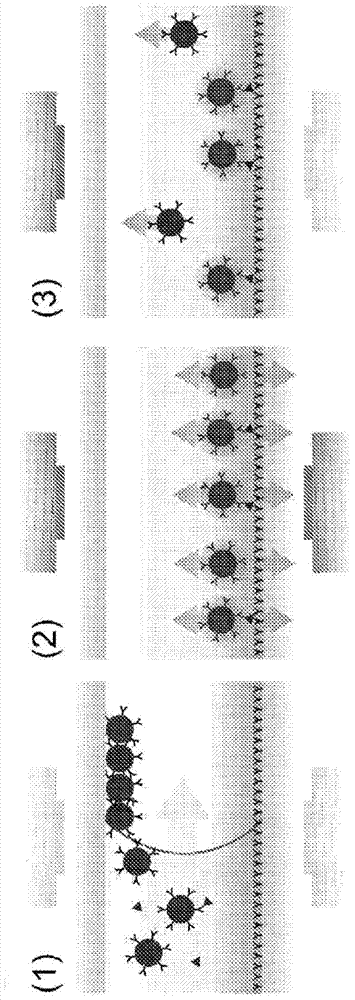
[0270] The effect of linker surface density on binding kinetics was tested in an assay format using streptavidin-coated 500 nm particles that bind to a single "target molecule" that is present in A dsDNA strand with biotin at one end and a Texas Red molecule attached to the other end (see Figure 9 , left panel, diamonds), which can be used to bind to the sensor surface coated with anti-Texas Red antibody. Subsequently, different amounts of non-functional dsDNA of the same length (but lacking the Texas Red molecule) were also bound to the particles, and the ability of the particles to bind to the sensor surface was compared.
[0271] In a further experiment, particles were loaded with 1999bp dsDNA. Figure 9 , middle panel, shows the relative amount of particle binding after loading with non-functional DNA, compared to particles with only one "functional" DNA molecule.
[0272] In further experiment...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com