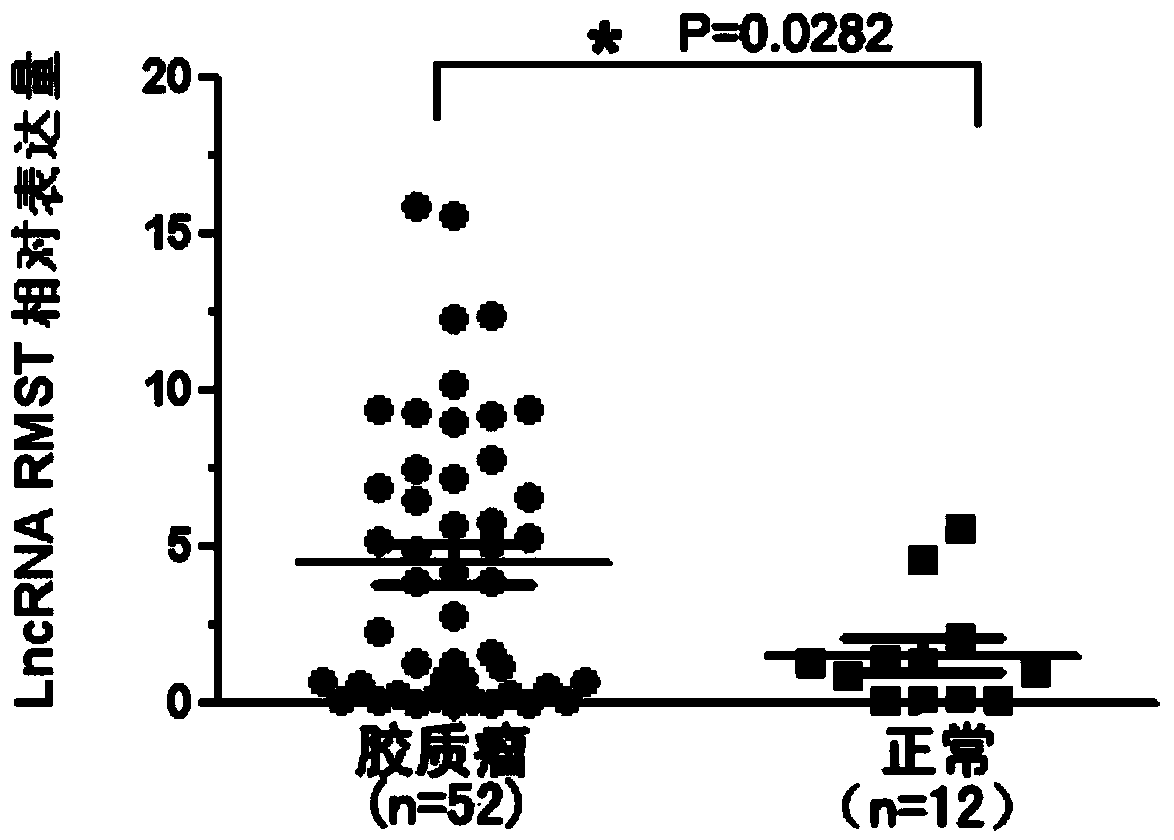
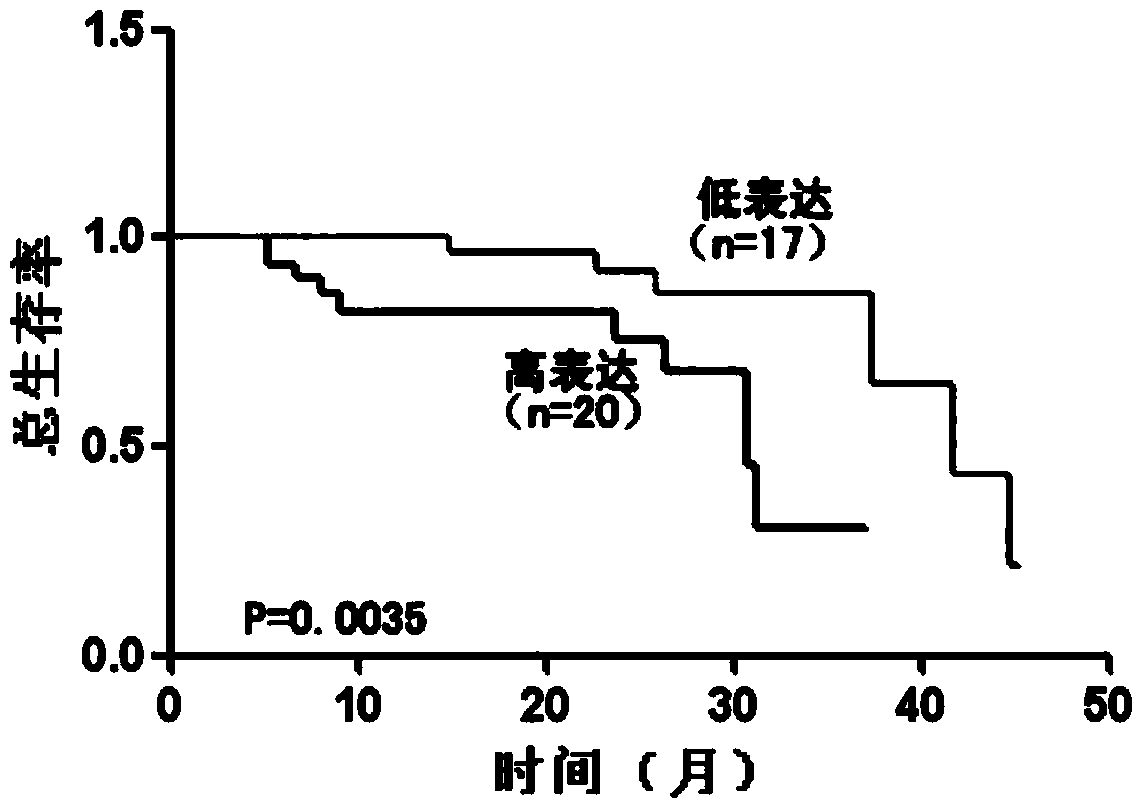
Application method of long no-coding RNA (Lnc RNA (Ribonucleic Acid)) RMST (Rhabdomyosarcoma 2 associated Transcript)
A long-chain non-coding and application method technology, which is applied in the application field of preparing prognostic reagents for glioma patients, can solve the problem of inability to adapt to the prognosis judgment of human gliomas, poor prognosis of malignant gliomas in glioma patients, no specificity Sexual indicators and other issues, to achieve far-reaching clinical significance and promotion effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1 Preparation of LncRNA RMST kit for the prognosis of glioma patients (50 reactions)
[0025] 1. RNA stabilization solution 50ml
[0026] 2. Isopropanol 100ml
[0027] 3. Chloroform 100ml
[0028] 4. Trizol 50ml
[0029] 5. Enzyme-free water 10ml
[0030] 6.1μM random reverse transcription primer 50ul
[0031] 7.5× reverse transcription buffer 200ml
[0032] 8.10mM base triphosphate deoxyribonucleotides 100ul
[0033] 9.40U / μl RNase inhibitor 500ul
[0034] 10.200U / μl MMLV reverse transcriptase 50ul
[0035] 11. Premix Ex Taq 50ul
[0036] 12.10μM Lnc RNA RMST specific primer 30ul
[0037] Forward primer 5′-AACTCCGTGTCCCTTGTG-3′
[0038] Reverse primer 5′-GGCAGTGGGTGACTGATC-3′
[0039] 13. 10μM U6snRNA specific primer 30ul
[0040] The forward primer is 5′-ATTGGAACGATACAGAGAAGATT-3′
[0041] The reverse primer was 5'-GGAACGCTTCACGAATTTG-3'.
Embodiment 2
[0042] Example 2 Detection of Lnc RNA RMST in Glioma Tissue
[0043] 1. Preservation of glioma tissue: collect the glioma tissue to be tested and store it in a cryopreservation tube filled with RNA stabilization solution, and put it in a -80°C refrigerator for later use.
[0044]2. Extraction of RNA in tissues: Take an appropriate amount of specimen, add liquid nitrogen to the mortar after baking at 180°C for 6-8 hours, grind the specimen, grind to powder, add 1ml Trizol mortar specimen to the mortar, and grind into After liquid, transfer to tube, add 200μl / ml Trizol in chloroform, shake by hand for 15-30s, place on ice for 5min, centrifuge at 12000g at 4°C for 15min; carefully take the upper aqueous phase into a new tube, add pre-cooled Mix with isopropanol 0.5ml / ml Trizol, put it in a refrigerator at -20°C for 20min, centrifuge at 12000g at 4°C for 10min; discard the supernatant, add 1-2ml of ethanol diluted with 75% DEPC water, mix well, centrifuge at 7500g at 4°C for 5min,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com