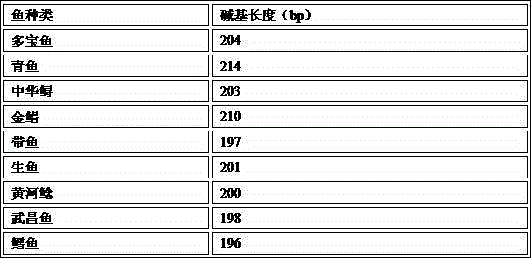
DNA Barcoding-based identification kit for 9 normal fishes
A fish and herring technology, applied in the field of fish identification, can solve the problems of being unable to screen and identify fish, time-consuming, and high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0016] 1. DNA extraction from fish:
[0017] Refer to the "Guidelines for Molecular Cloning Tests" with slight modifications: (1) Take 10 mg of muscle tissue from each sample, and use ophthalmic anatomical scissors to cut it into pieces as much as possible. (2) Carefully pack the extracted sample into a 1.5ml autoclaved centrifuge tube, and then add 600ul of SET (lysate). (3) Add 10ul of Prok (20) to each EP tube, then add 15ul of SDS, and mix well. (4) Digest overnight in a 55°C water bath to fully digest the muscle tissue. (5) Add an equal volume of about 600u of Tris-saturated phenol to each EP tube, shake and mix for 15 minutes, and then centrifuge at 1200 rpm for 10 minutes. (6) Take out the supernatant, put it into a new EP tube, add an equal volume of Tris-saturated phenol, shake and mix for 15 minutes, and centrifuge at 12,000 rpm for 10 minutes. (7) Take the supernatant, add an equal volume of phenol: chloroform: isoamyl alcohol (25:24:1) and mix for 10 minutes, th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com