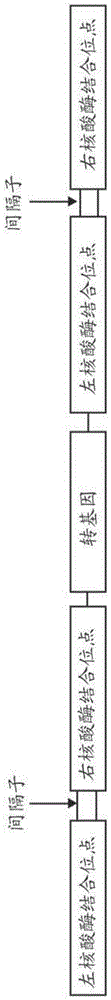
Methods and compositions for nuclease-mediated targeted integration of transgenes
A nuclease and transgenic technology, applied in the field of genome engineering, can solve problems such as the lack of HDR process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0205] Example 1: Materials and methods
[0206] Cell Growth, Transfection, and ZFN / TALEN Assays
[0207] K562 (ATCC CCL-243) transfection uses Amaxa solution V and T-016 program; CHO-K1 (ATCC CCl-61) transfection uses Amaxa solution T and U-023 program. All transfections contained 10 6cells and the following plasmids: AAVS1, 3 μg of 2A-linked ZFN and donor; IL2Rγ and CCR5, 2 μg of each unlinked ZFN plasmid and 10 μg of donor plasmid; GS, FUT8, 3 μg of 2A-linked ZFN and 10 μg of donor plasmid. See, US Patent No. 8,110,379 and US Patent Publication Nos. 20100129869 and 20090042250 and DeKelver et al. (2010) Genome Res. 20(8):1133-1142 for additional details on designs involving ZFNs; Liu et al. (2009) Biotechnol.Bioeng.106(1):97-105; Malphettes et al. (2010) Biotechnology and Bioengineering 106(5):774-83; Perez et al. (2008) Nature Biotech.26(7):808-816; Urnov et al. (2005) Nature 435(7042):646-651.
[0208] The FUT8 TALE nuclease pair (SBS 101082 and SBS 101086) directly ...
Embodiment 2
[0222] Example 2: Targeted integration following in vivo cleavage of a double-stranded donor
[0223] A.AASV1
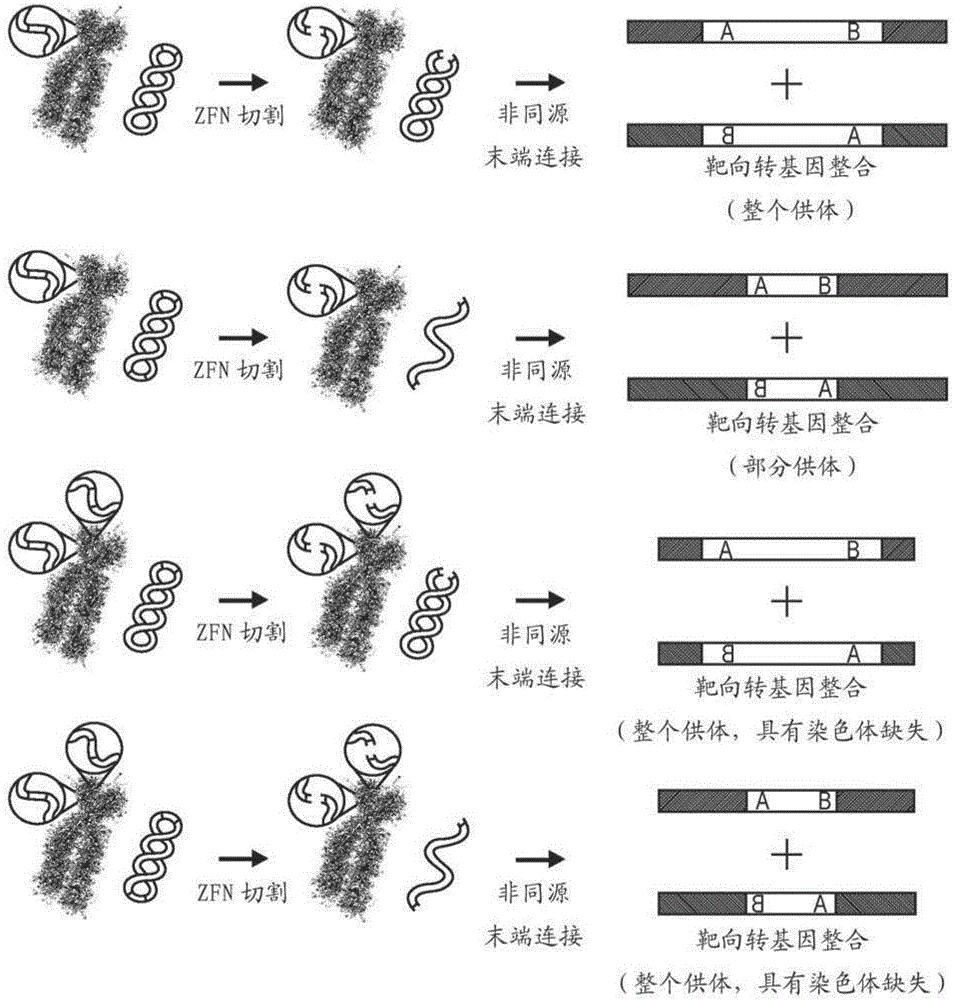
[0224] To test whether in vivo transgene cleavage with the same ZFN that cleaves the genomic target site would simultaneously undergo donor and chromosomal cleavage, thereby minimizing the transgene's vulnerability to degradation, an AASV1-targeted ZFN was utilized with K562 cells were transfected with the donor plasmid for in vivo cleavage of the AASV1 ZFN target site of the donor plasmid. Briefly, as described in Example 1, we cloned the recognition site for the well-characterized and highly active AASV1 ZFN into a donor plasmid containing an autonomous GFP expression cassette but lacking homology to the AASV1 locus. See, US Patent No. 8,110,379 and DeKelver et al. (2010) Genome Res. 20(8):1133-1142. Donor plasmids (with or without ZFN target sites) were co-transfected into K562 cells with a second plasmid encoding AASV1 ZFN.
[0225] Insertion into the chromoso...
Embodiment 3
[0236] Example 3: In vivo and in vitro cleavage
[0237] To determine whether in vivo cleavage was necessary to support the observed levels of target gene insertion, we utilized direct comparisons of targeted integration using EcoRV in vivo cleaved donors and in vitro cleaved donors, as described in Example 1.
[0238] As shown in Figure 2, although integration of the precut donor plasmid was occasionally detectable, it was significantly less efficient than the in vivo cleaved donor ( Figure 2A , compare lanes 2 / 3, 6 / 7, 10 / 11, 14 / 15, 18 / 19 and 22 / 23). Furthermore, the use of pre-cut donor DNA prior to chromosome capture showed an increased range of adapter PCR sizes consistent with increased levels of donor DNA degradation (see, e.g., Figure 2A , lane 22).
[0239] To determine whether targeted integration could be stimulated through the use of two different nucleases (ZFNs), we used the GS ZFN pair (Example 1) to cleave the chromosome of CHO-K1 cells and the AASV1 ZFN pai...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Melting temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com