Universal primer sequence for detecting human coronavirus infection and detection method
A human coronavirus and universal primer technology, applied in the field of qualitative detection of human coronavirus infection, can solve the problems of easy pollution, expensive detection reagents, time-consuming and labor-intensive problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
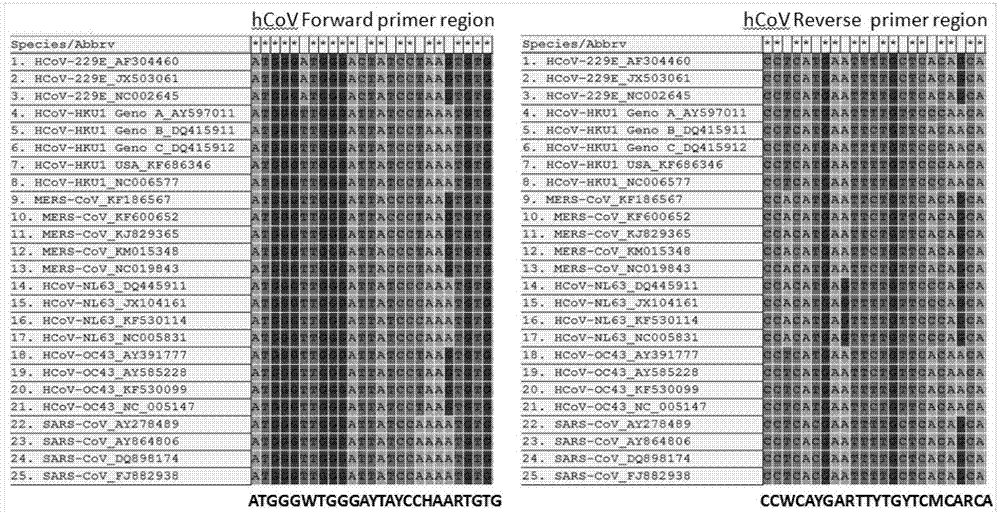
[0021] A universal primer sequence for detecting human coronavirus infection, its upstream primer nucleotide sequence is shown in SEQ ID NO: 1, that is: hCoV-For: ATGGGWTGGGAYTAYCCHAARTGTG; its downstream primer nucleotide sequence is as SEQ ID NO: 2, namely: hCoV-Rev: TGYTGKGARCARAAYTCRTGWGG.
[0022] A detection method for detecting human coronavirus infection with a universal primer sequence, comprising:
[0023] (1) Universal (degenerate) primer design for human coronavirus detection
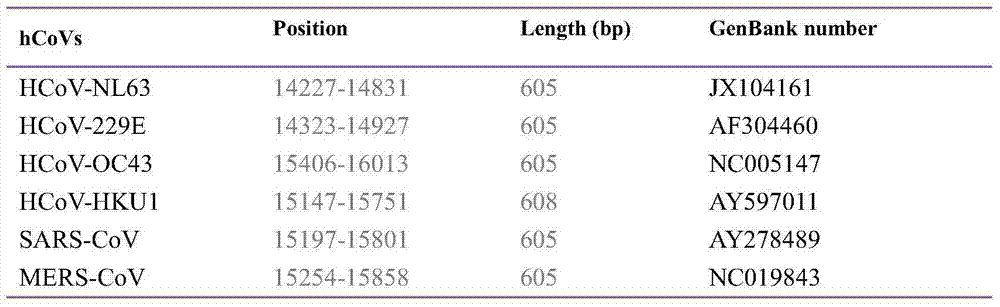
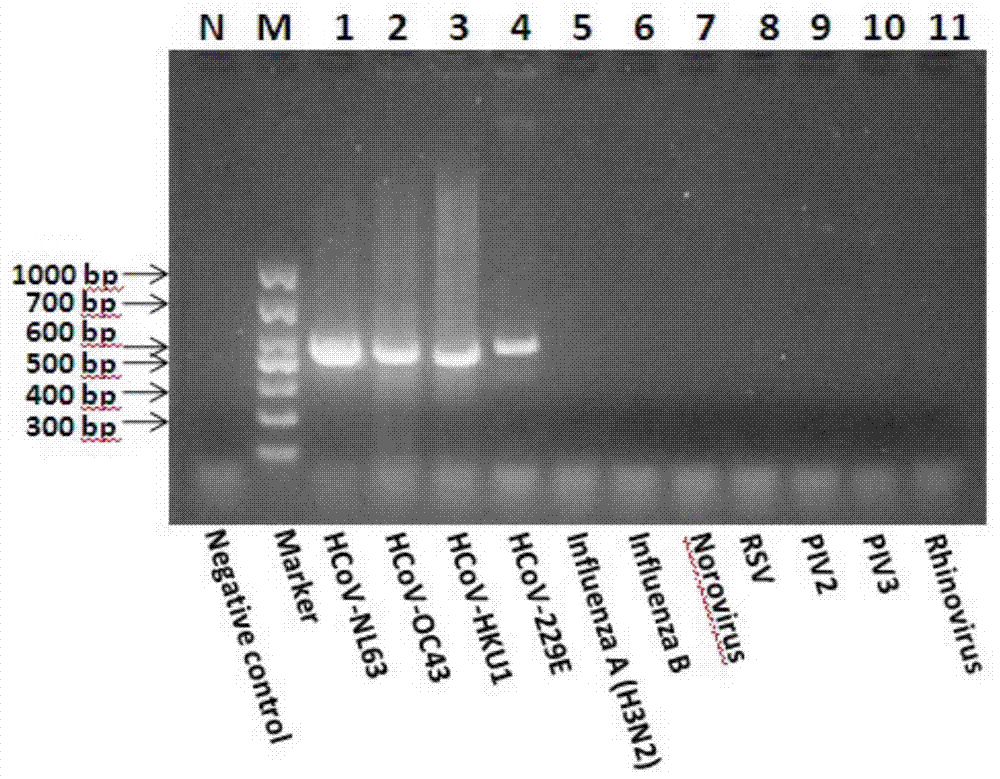
[0024]The six human coronaviruses registered on GenBank include human coronavirus 229E (Human coronavirus 229E, HCoV-229E), HCoV-OC43, SARS-CoV (Severe acute respiratory syndrome coronavirus), HCoV-NL63, HCoV-HKU1 and MERS -Comparative analysis of the whole genome nucleic acid sequence of CoV (Middle East respiratory syndrome coronavirus), a pair of degenerate primers hCoV-For and hCoV-Rev were designed in the genome polymerase gene coding region (region 1b) for the detection of human coron...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com