Reagent and method for detecting ninth exon mutation of CALR gene
A technology of exons and reagents, applied in the fields of life sciences and biology, can solve the problems of high cost of reagents, inapplicability of daily screening, inapplicability of deletion and insertion mutation detection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
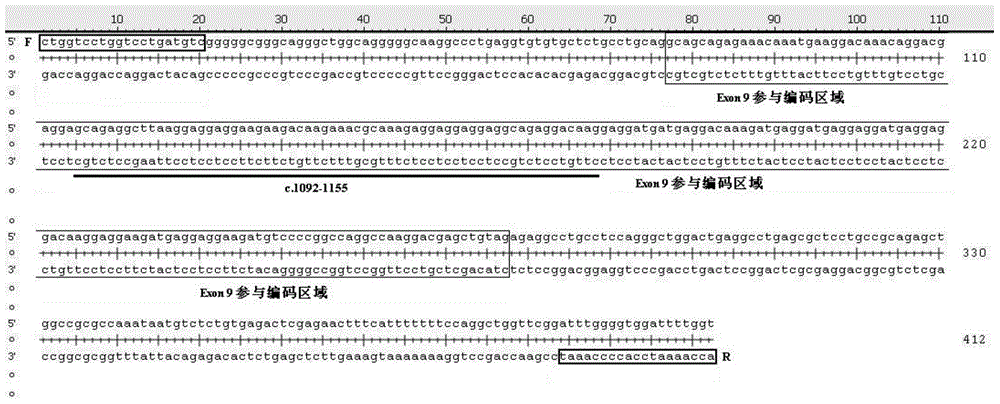
[0032] The present invention is used to detect the amplification primer of the 9th exon mutation of CALR gene in the sample as shown in the following table:
[0033]
[0034]
Embodiment 2
[0036] Blood DNA extraction: TIANamp Genomic DNA Kit Blood / Cell / Tissue Genomic DNA Extraction Kit (Tiangen Biochemical Technology (Beijing) Co., Ltd.) was used. Take 300μl of the whole blood sample to be tested and add it to a 1.5ml centrifuge tube, add 900ul red blood cell lysate and mix it upside down, centrifuge at 10000rpm for 1min, discard the supernatant; add 200uL buffer GA, shake until thoroughly mixed; add 20μl proteinase K solution, mix Mix well; add 200μl buffer GB, mix thoroughly by inversion, and place at 70°C for 10min. Add 200 μl of absolute ethanol, mix well, add the solution to an adsorption column CB3 (the adsorption column is placed in a collection tube), centrifuge at 12000rpm for 30s, discard the waste liquid; add 500μl buffer GD to CB3, centrifuge at 12000rpm for 30s, discard Waste liquid; add 600μl rinse solution PW to CB3, centrifuge at 12000rpm for 30s, discard waste liquid; repeat once. Put CB3 back into the collection tube and centrifuge at 12000rpm...
Embodiment 3
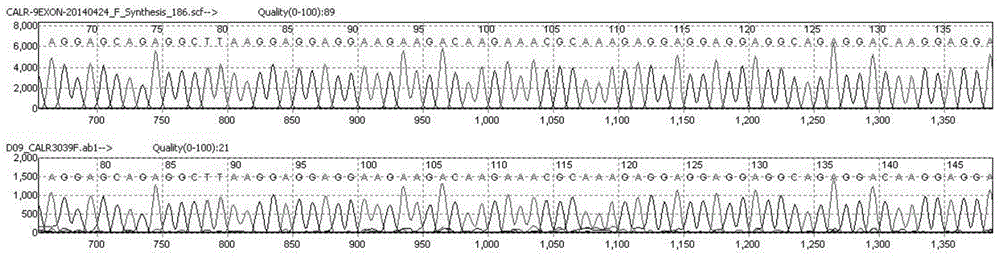
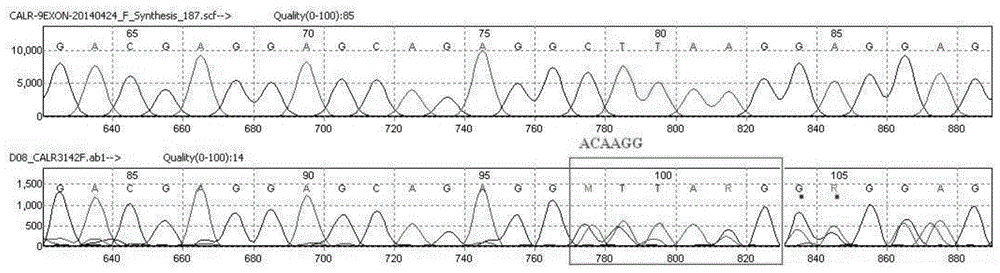
[0039] PCR amplification: Configure the PCR amplification system according to the following reagents and reagent volumes, including 0.3ul each of primers CALR-F (10μM) and CALR-R (10μM); 2×KOD Buffer 10ul, KOD enzyme (1U / ul) 0.3ul , d NTP (2mM) 2ul (Toyobo (Shanghai) Biotechnology Co., Ltd.); add 6.1ul of deionized water; finally add 1ul of DNA template. The amplification program is as follows: 95°C for 10 minutes; 98°C for 10s, 62°C for 30s (1°C drop per cycle), 68°C for 30s, 10 cycles; 98°C for 10s, 54°C for 30s, 68°C for 30s, 20 cycles; 68°C ℃ 2min.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com