Method for recognizing protein network compound based on semantic density
A protein network and semantic density technology, applied in the field of data mining, can solve the problems of increasing the time complexity of the algorithm, and the modularity has a decomposition limit.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
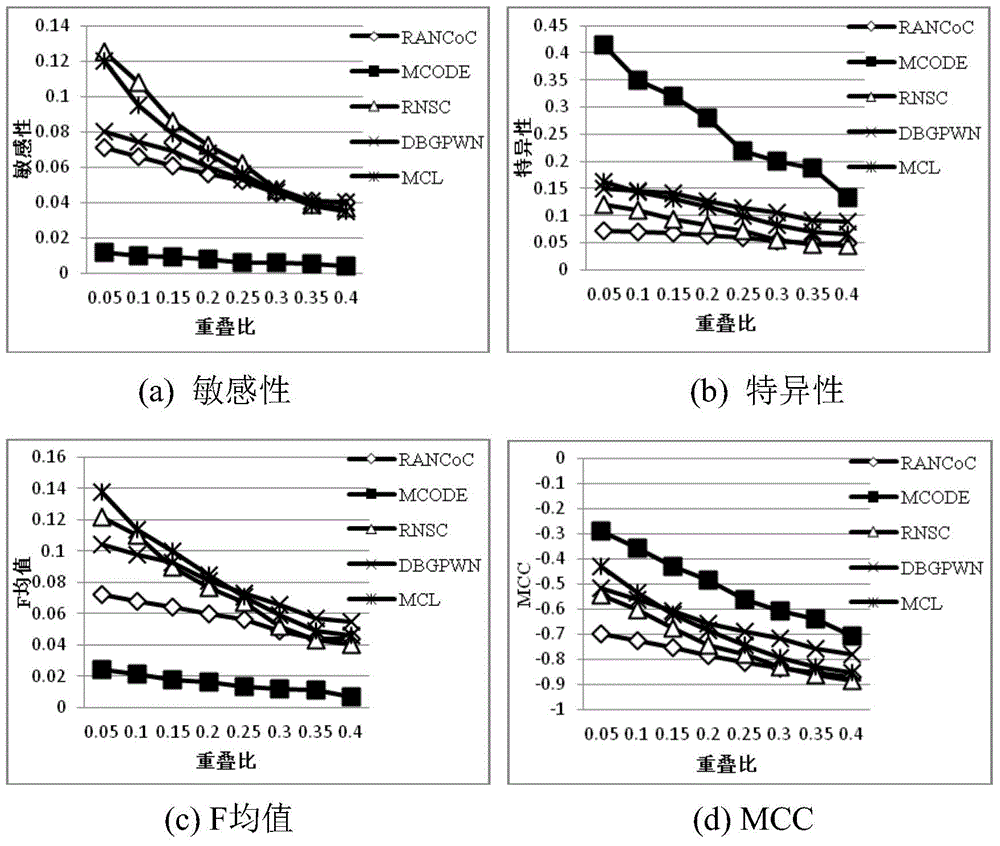
Method used
Image
Examples
Embodiment Construction
[0043] The present invention will be described in detail below in conjunction with the accompanying drawings and specific embodiments.
[0044] Related concepts and definitions involved in a protein network complex recognition method based on semantic density of the present invention are as follows:
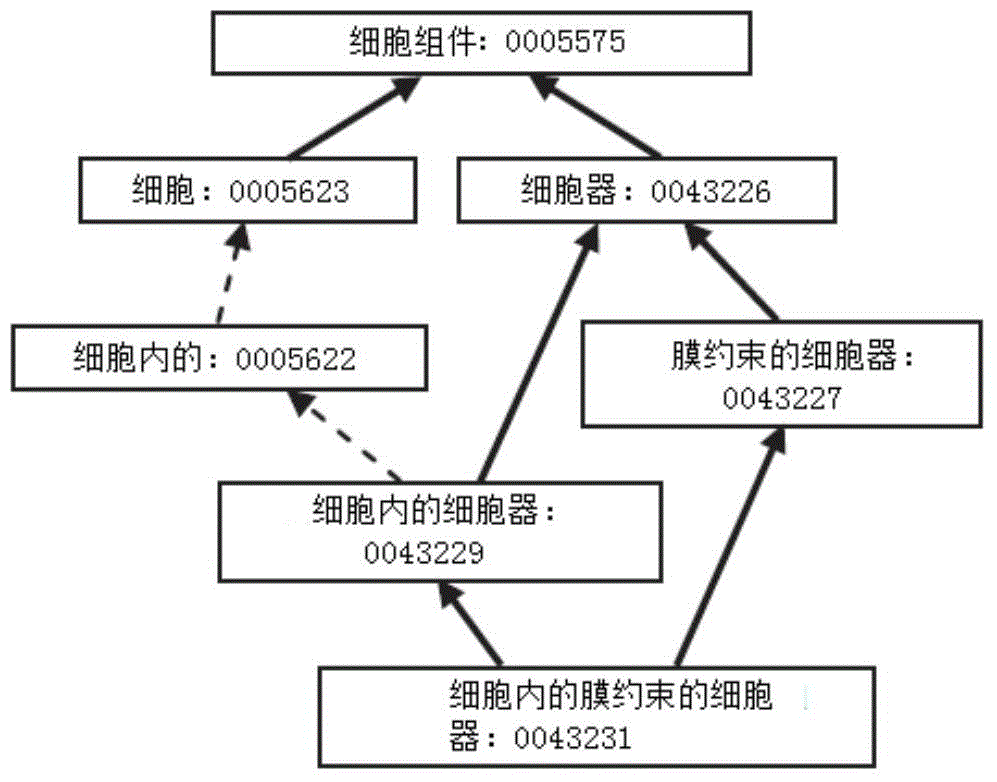
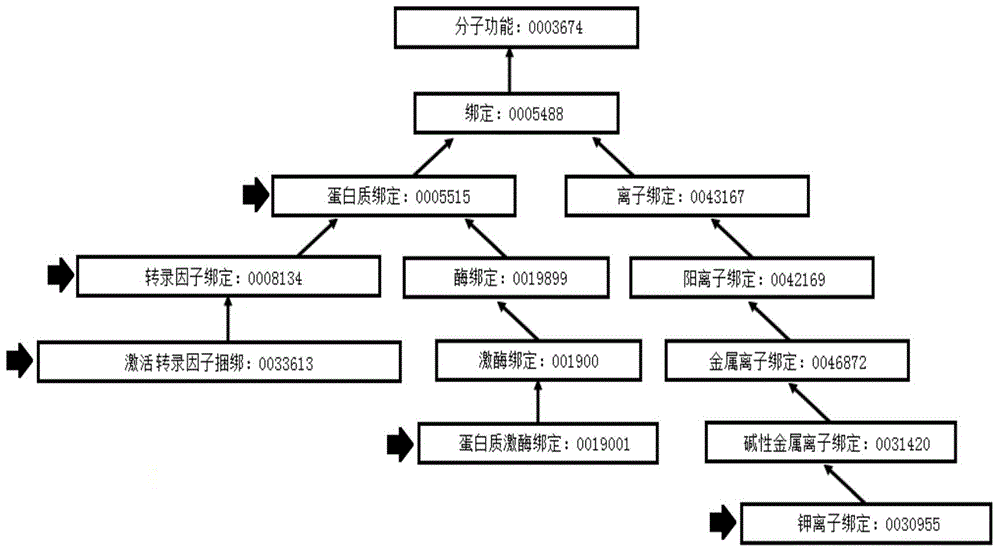
[0045] Gene Ontology database (Gene Ontology, GO) is a large-scale biological information database, and its goal is to unify the expression of all biological genes and protein properties. GO collects biological attributes as well as biochemical attributes to describe the function of the corresponding protein or its location in the cell. All attributes are divided into three categories: biological process, molecular function, and cellular component, abbreviated as P, F, and C, respectively. As an ontology, GO contains two semantic relations, namely "is_a" and "part_of". The directed acyclic graph (DAG) in the data structure is usually used to represent GO. The nodes in the graph...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com