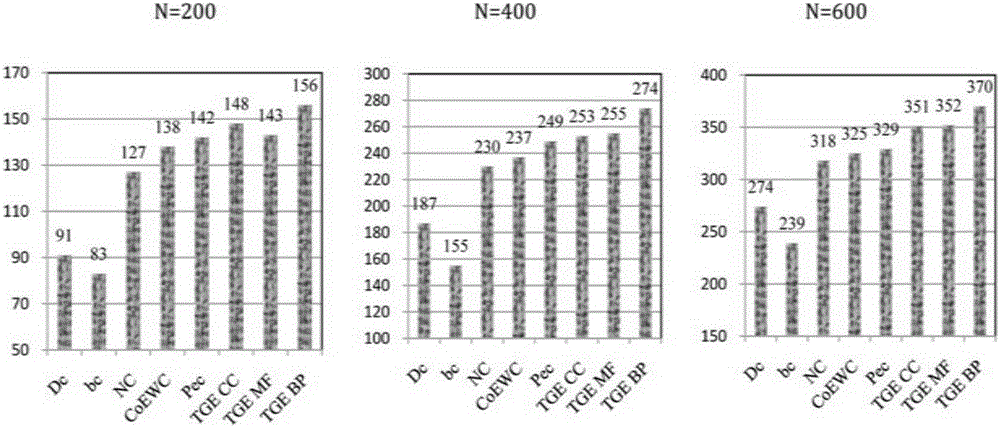
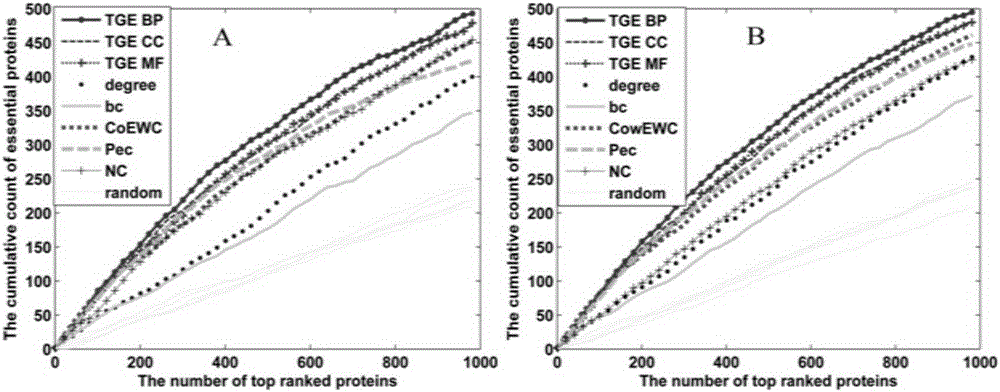
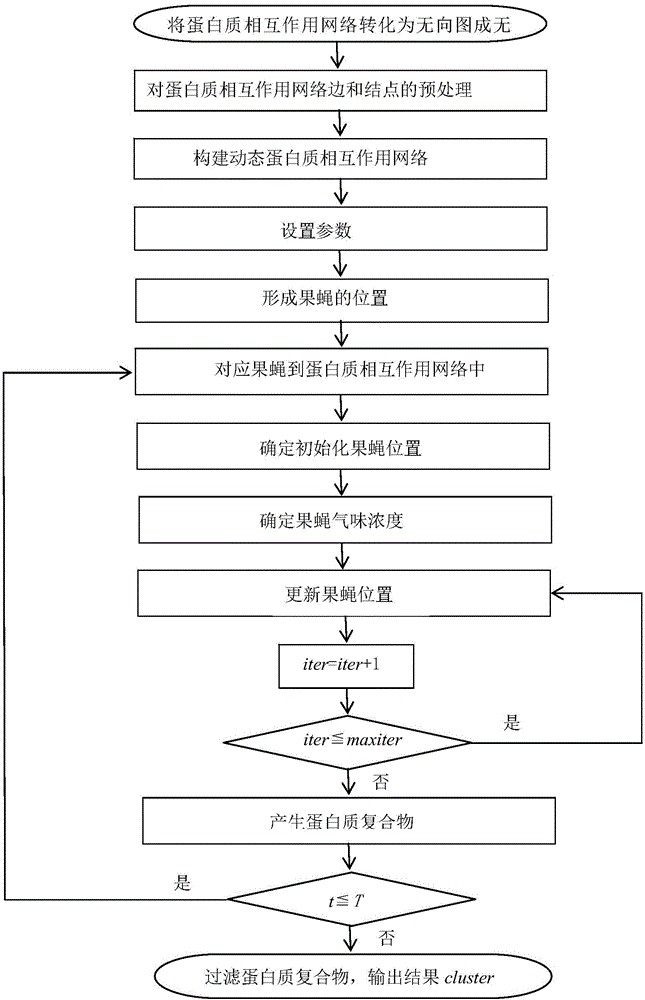
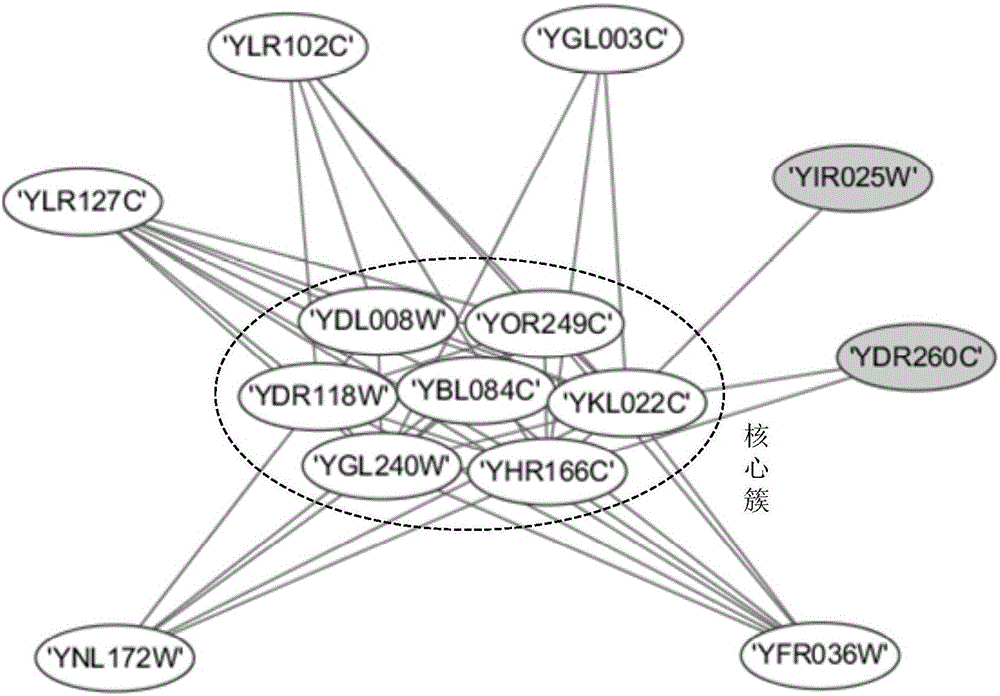
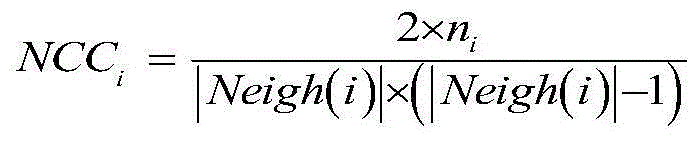Patents
Literature
30 results about "Protein protein interaction network" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Method for identifying protein functions based on protein-protein interaction network and network topological structure features
InactiveCN105138866ARobustSignificant predictive advantageSpecial data processing applicationsNODALData set
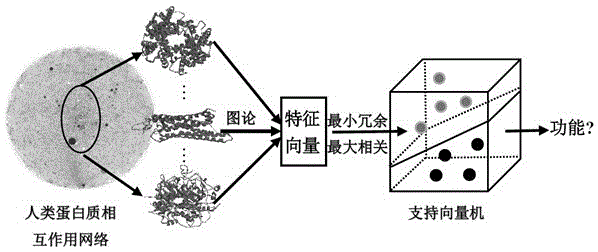
The invention discloses a method for identifying protein functions based on a protein-protein interaction network and network topological structure features. Firstly, a node and side-weighted protein-protection interaction network is established, wherein the node represents protein while the edge represents the interaction; then the nodes and the sides in the network are weighted by protein first-grade structural description and protein-protein interaction trust scoring; protection functional annotation data is collected to establish a data set, and a new protein with overall and local information network topological structure features is provided based on a graph theory; and finally, the protein functions are predicated by choosing features through adopting a minimum-redundancy maximum-correlation method and by modeling through a support vector machine. The protein function predication method is greatly better than the prior art, and has robustness on sequence similarity and sampling; and meanwhile, information of three-dimensional structure and the like of protein is not required, so that the method is simple, rapid, accurate and efficient, and the method is expected to be applied in the research fields of proteomics and the like.
Owner:SYSU CMU SHUNDE INT JOINT RES INST +2
Method for identifying key proteins in protein-protein interaction network
ActiveCN105279397AImprove recognition accuracySolve the costSpecial data processing applicationsProtein insertionProtein protein interaction network
The invention discloses a method for identifying key proteins in a protein-protein interaction network. According to the method, an undirected graph G is constructed according to the protein-protein interaction data, and the edge clustering coefficient of the graph is calculated. Compared with the prior art, the method provided by the invention has the advantages of combining the gene expression profile data and the gene function annotation information data on the basis of considering the topological structure characteristics of the protein-protein interaction network, and integrating three groups of data to predict the key proteins, so that the influence caused by the data noise of a single data source on the prediction correctness can be effectively decreased, and the key proteins in the network can be predicted through the key protein characteristics embodied by three types of data, such as the edge clustering coefficient in the protein-protein interaction network, the Pearson's correlation coefficient of the gene expression value and the gene function similarity index. According to the method, the identification correctness of the key proteins in the protein-protein interaction network can be remarkably improved, and abundant key proteins can be predicted once, so that the problem that the biological experiment method is high in cost and time-consuming is solved.
Owner:EAST CHINA JIAOTONG UNIVERSITY
A method of identifying protein compounds by using a fruit fly optimization method
InactiveCN105868582AImprove accuracyRealistically simulate dynamicsProteomicsGenomicsProtein protein interaction networkProtein-protein complex
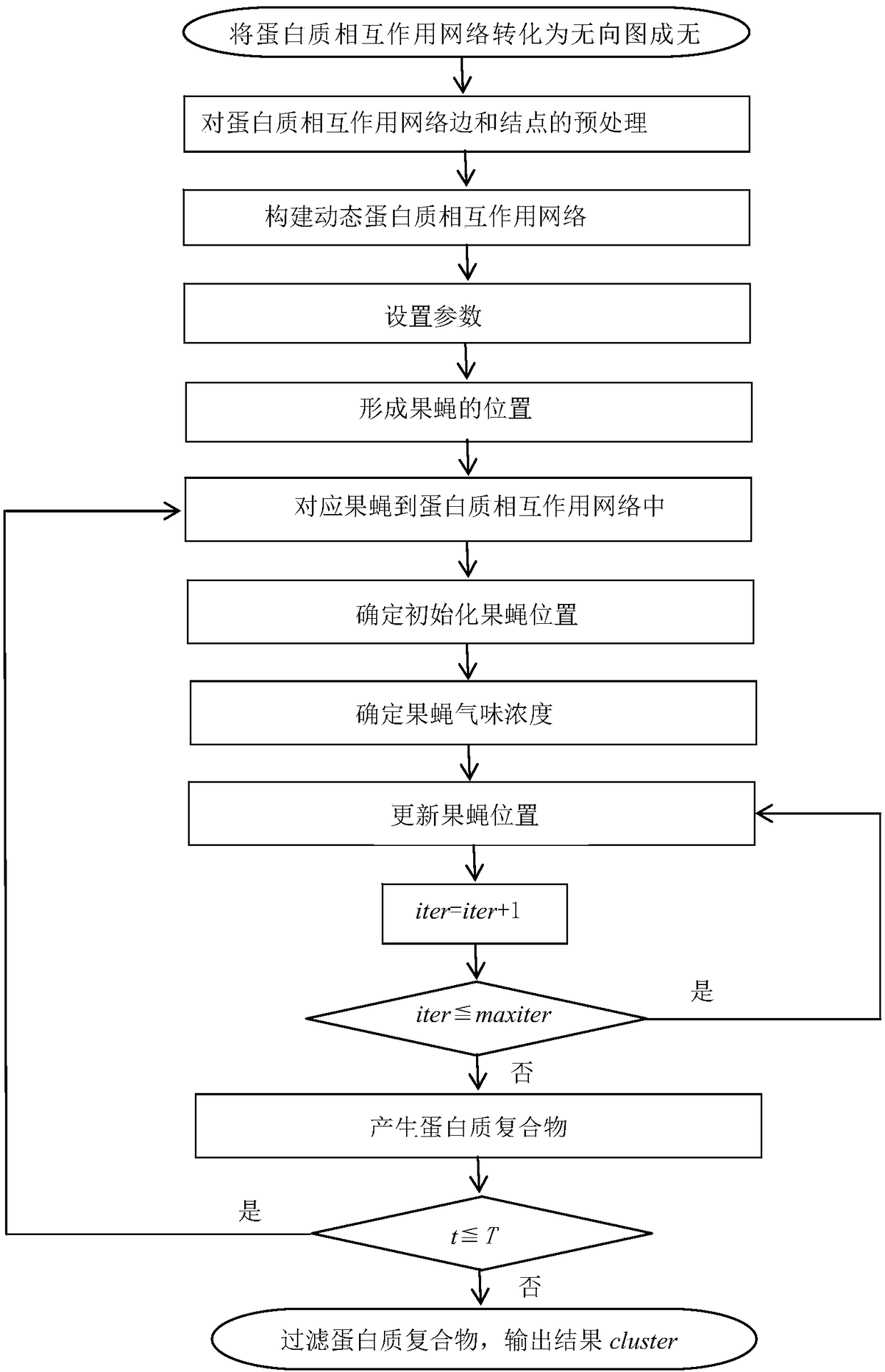
The invention provides a method of identifying protein compounds by using a fruit fly optimization method. The method comprises the steps of converting a protein-protein interaction network into a undirected graph, performing pretreatment on the edges and nodes of the protein-protein interaction network, establishing a dynamic protein-protein interaction network, setting parameters, forming fruit fly positions, matching fruit flies with the protein-protein interaction network, determining initialization fruit fly positions, determining the fruit fly odor concentration, updating the fruit fly positions, generating a protein compound, and filtering the protein compound. The method gives full consideration to the dynamic nature of the protein network, the protein compound inner core-attachment structure and the locality and wholeness of the protein-protein interaction network and can identify protein compounds accurately. The results of simulation experiments show that the performance of the indexes such as the accuracy and the recall ratio are excellent. Compared with other clustering methods, the method, based on the characteristics of the protein network and the protein compounds, realizes the protein compound identification process and improves the protein compound identification accuracy.
Owner:SHAANXI NORMAL UNIV
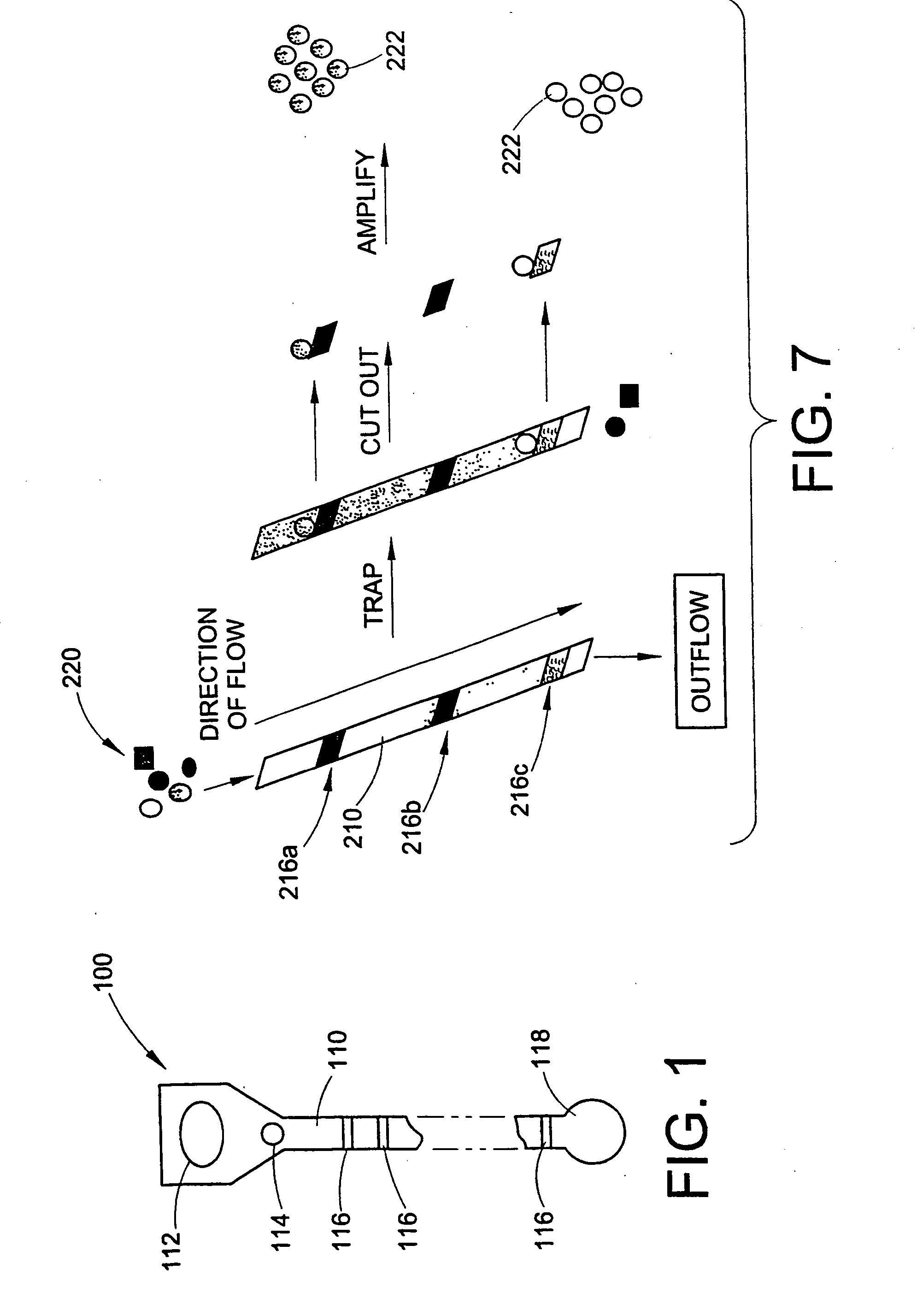
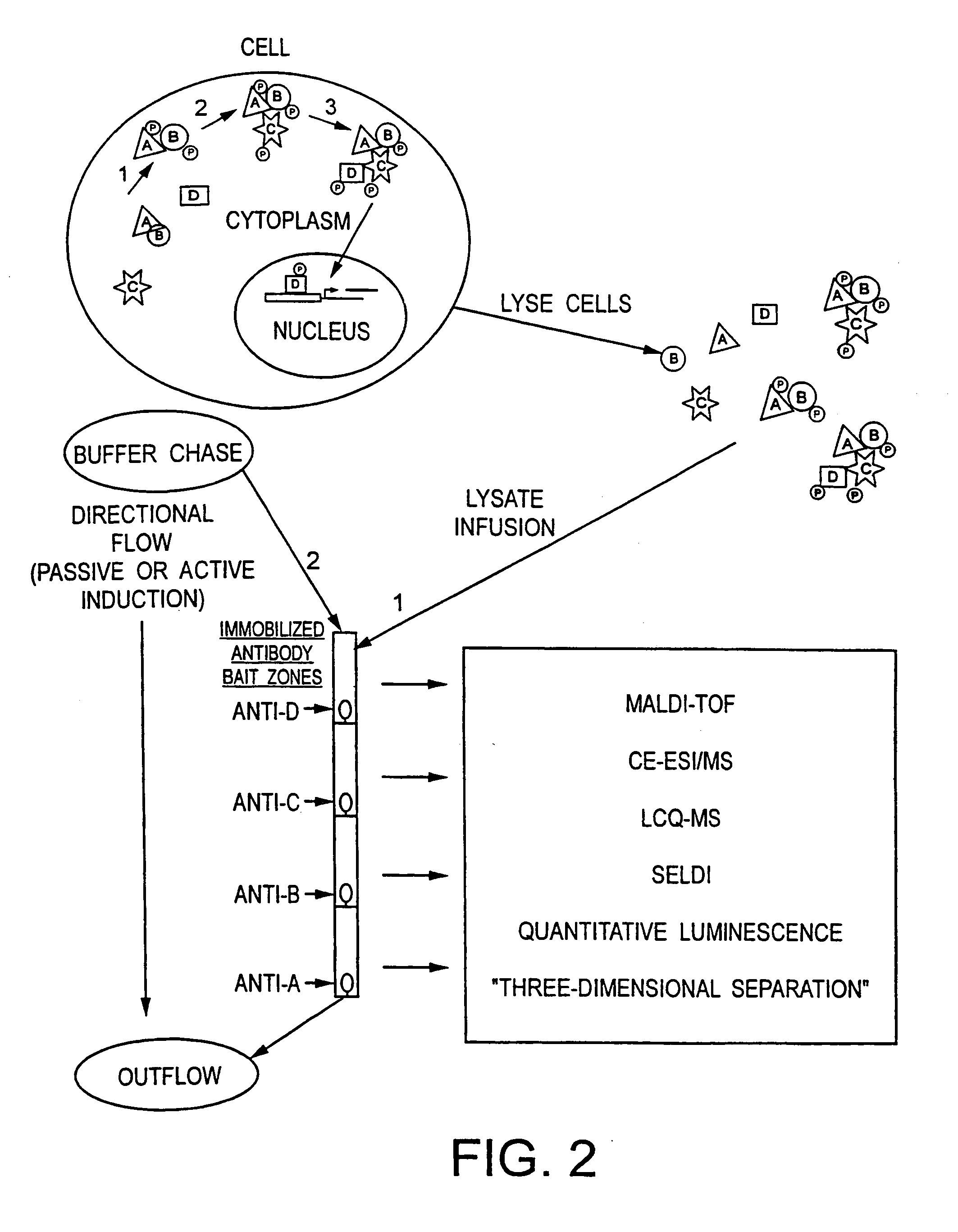
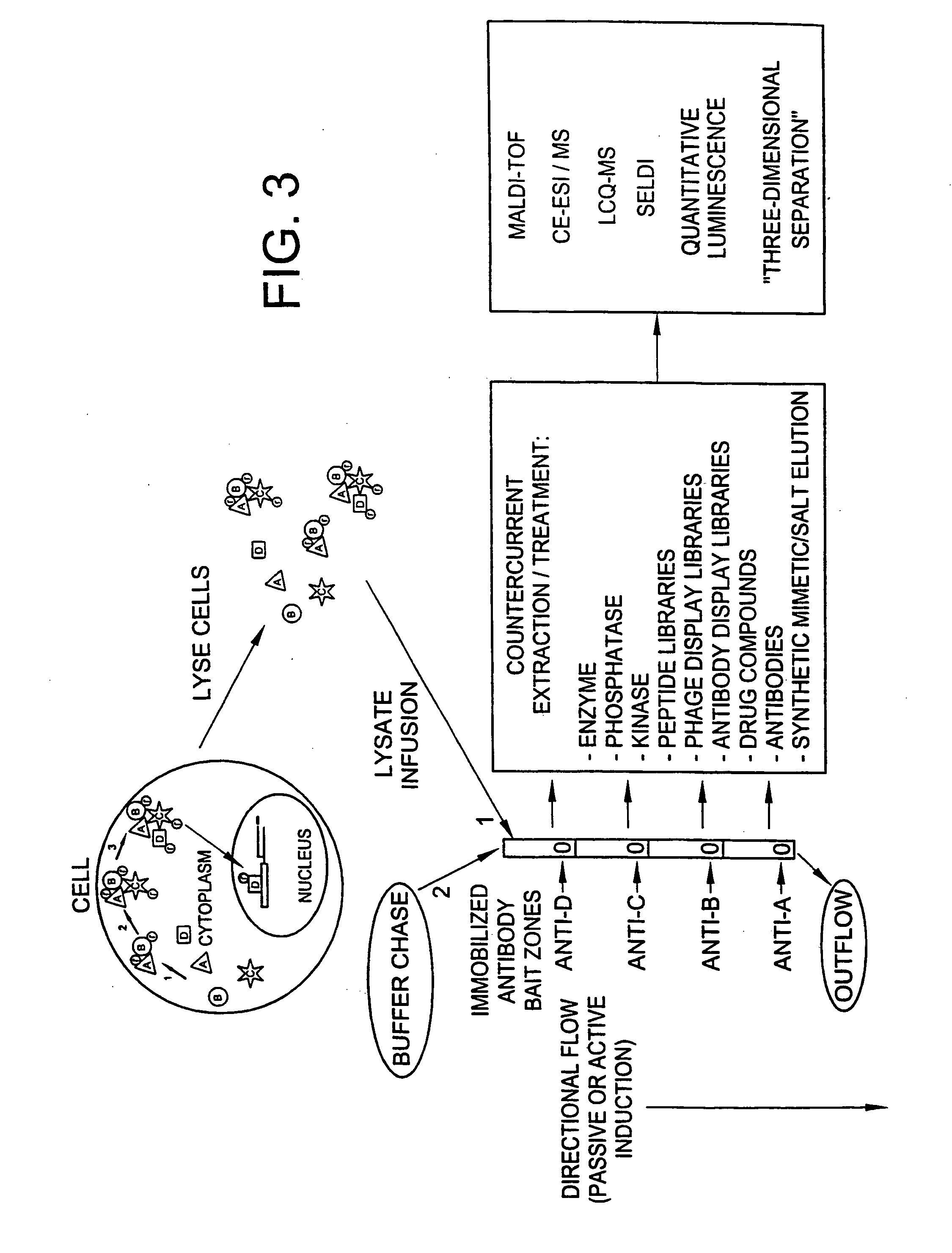
Method and apparatus for signal transduction pathway profiling
ActiveUS20070105157A1Bioreactor/fermenter combinationsBiological substance pretreatmentsAnalyteProtein protein interaction network
An assay device for determining the presence of analytes in a cell lysate comprises a porous support member and a plurality of binding reagents arranged and immobilized at multiple reaction sites on the support member. The binding reagents are selected and arranged to assess the status of a selected cellular signal transduction pathway / protein-protein interactive network. In a further aspect, a method for assessing the status of a signal transduction pathway comprises generating a lysate of cells, the lysate retaining one or more pathway molecules present in one or more states and the pathway molecules reflecting signal transduction events taking place in the cells. The method further includes applying the lysate to an immobilized series of binding reagents which can discriminate the pathway molecules and their states. Binding events between the pathway molecules and the binding reagents are identified and the state of the selected signal pathway is determined.
Owner:INSTANT MEDICAL DIAGNOSTICS
Method for recognizing protein network compound based on semantic density
InactiveCN104992078AReduce time complexityShort timeSpecial data processing applicationsData setGene ontology
The invention discloses a method for recognizing a protein network compound based on a semantic density, which is specifically implemented at the steps that all the protein properties in a network data set are searched in a gene ontology base GO as for a protein-protein interaction network data set without a weight; based on searching results, similarity of connected proteins in the network data set is calculated by a semantic similarity calculation method based on gene ontology; according to obtained similarity results, the given protein-protein interaction network data set is converted into an undirected network data set with a weight, wherein nodes represent the proteins, borders stand for interactions among the proteins, and the similarity among the proteins is the weight of the border; and the protein compound can be recognized from a protein-protein interaction network, wherein recognition accuracy is high and time complexity is low.
Owner:XIAN UNIV OF TECH
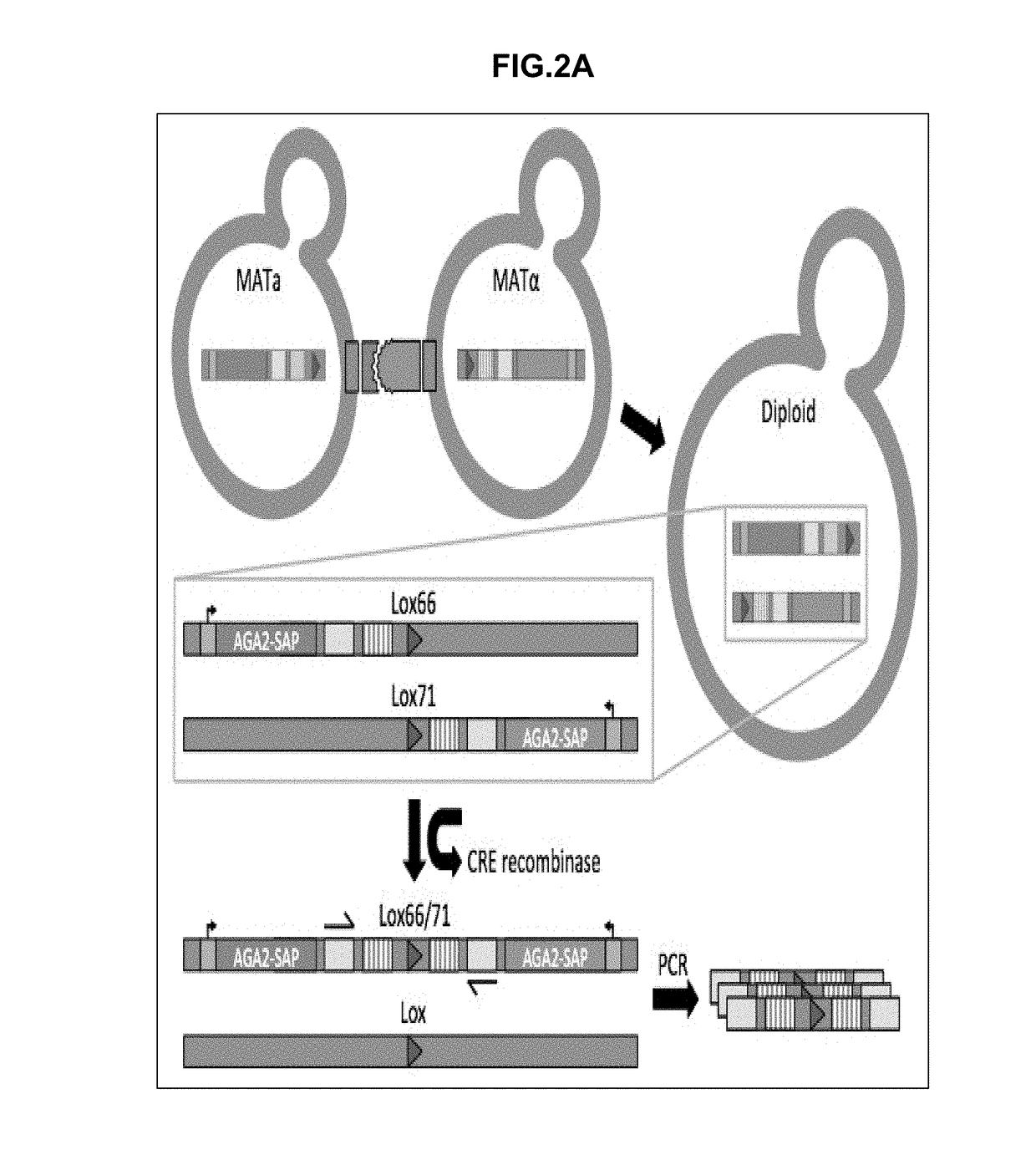
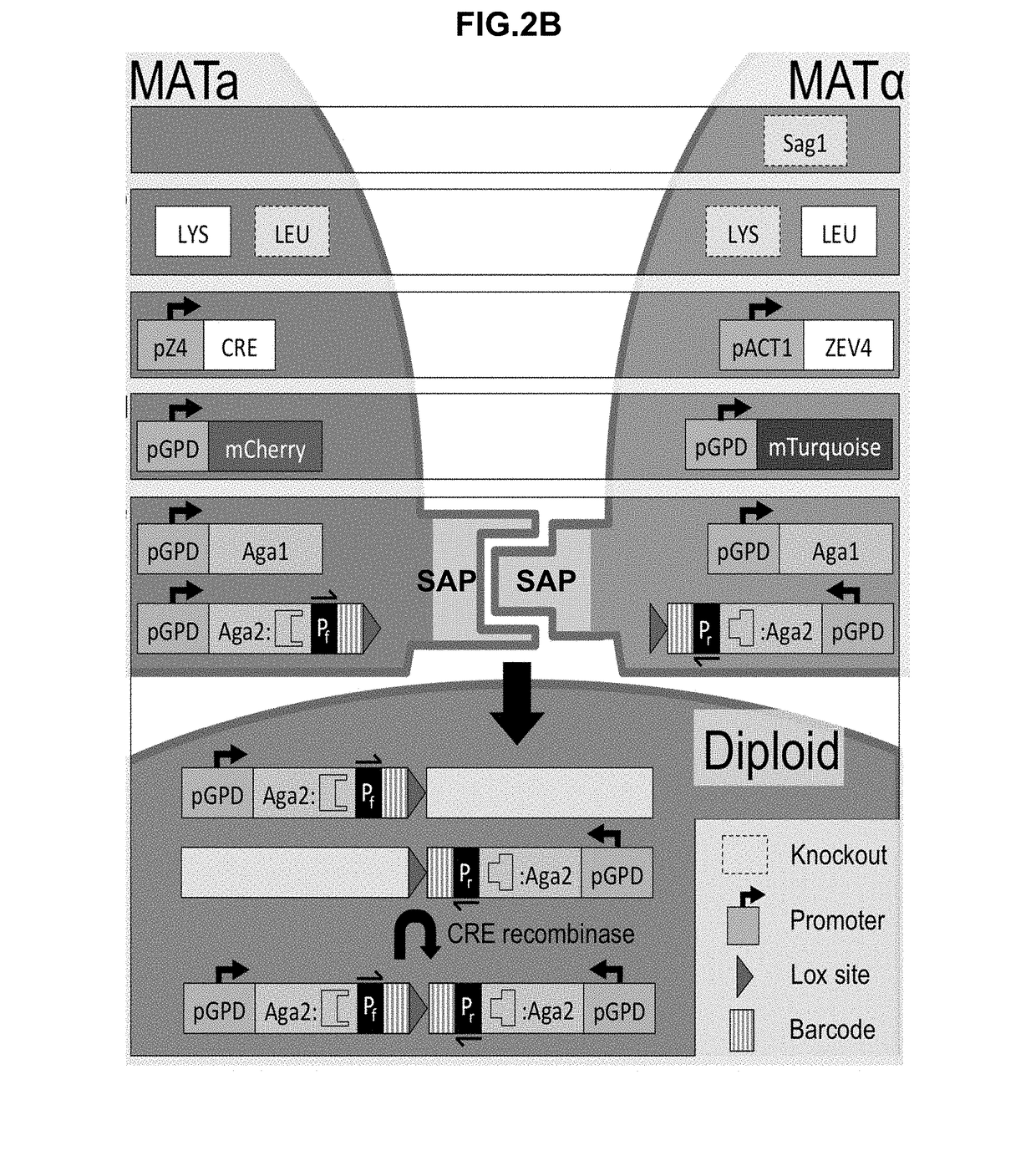
Synthetic yeast agglutination
InactiveUS20170205421A1High throughput screeningHydrolasesMicrobiological testing/measurementProtein protein interaction networkAgglutination
The present invention provides methods and compositions that can be applied to screening protein-protein interaction networks, screening drug candidates that modulate protein-protein interactions for on- and off-target effects, detecting extracellular targets for which no native S. cerevisiae receptor exists, and re-engineering yeast agglutination in order to answer biological questions about yeast speciation and ecological dynamics
Owner:UNIV OF WASHINGTON
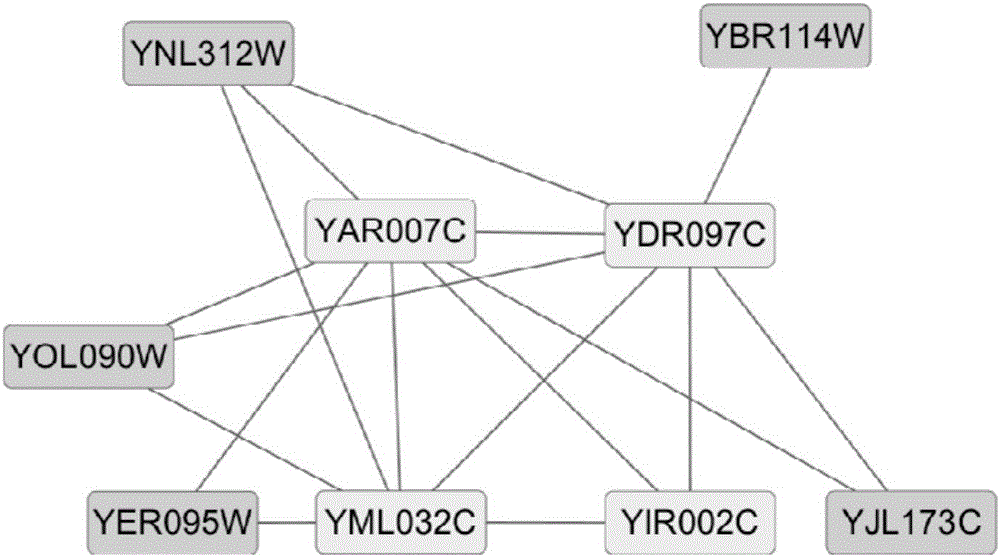
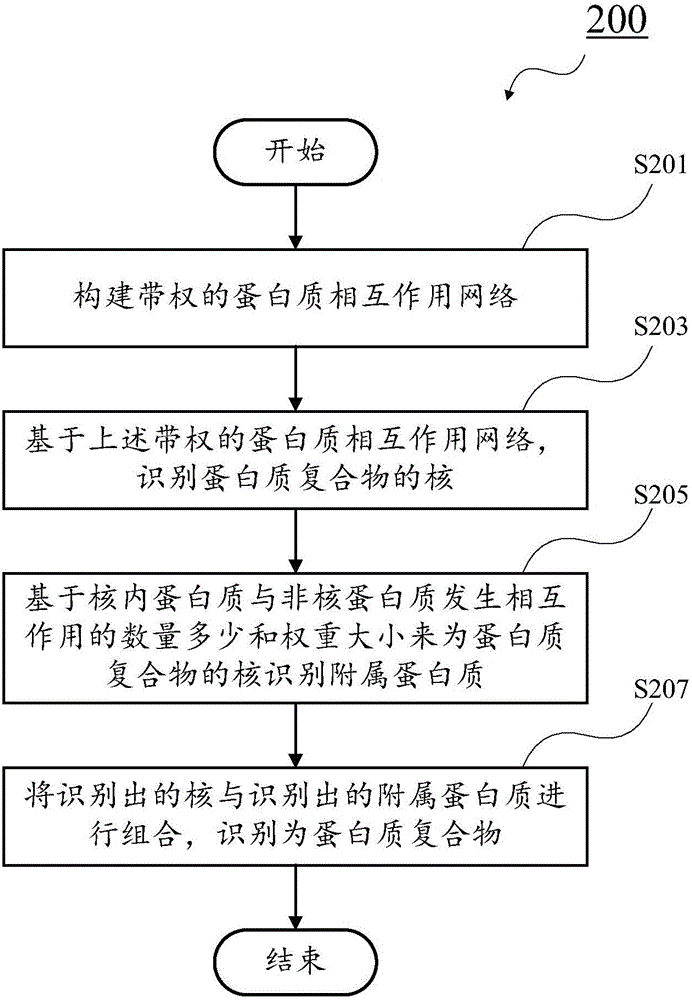
Recognition method of protein complexes
The invention discloses a recognition method of protein complexes. The method comprises following steps: constructing a weighted protein-protein interaction network, wherein weight value represents similarity between gene representing modes corresponding to proteins with codes interacting each other; recognizing nucleuses of protein complexes based on the weighted protein-protein interaction network; recognizing accessory proteins for the nucleuses of protein complexes based on the number of nuclear proteins interacting with non-nuclear proteins and weight; and combining recognized nucleuses and recognized accessory proteins to recognize the protein complexes. The method does not take internal structures of protein complexes into consideration and utilizes the feature of code interaction with high protein gene expression. Therefore, the method is capable of recognizing protein complexes with overlapping structures and the recognized protein complexes reflect actual internal structures of protein complexes.
Owner:HENAN UNIV OF URBAN CONSTR
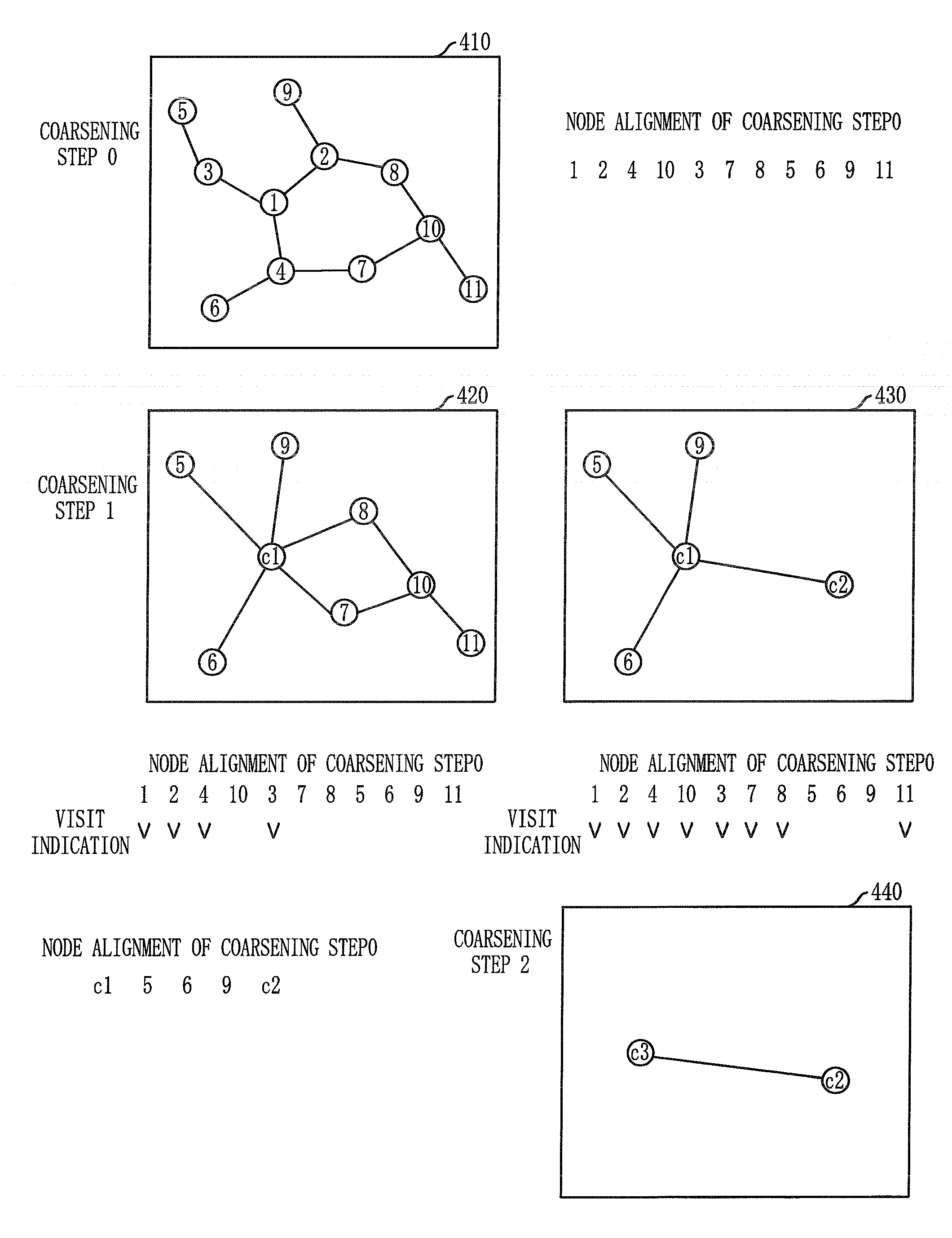
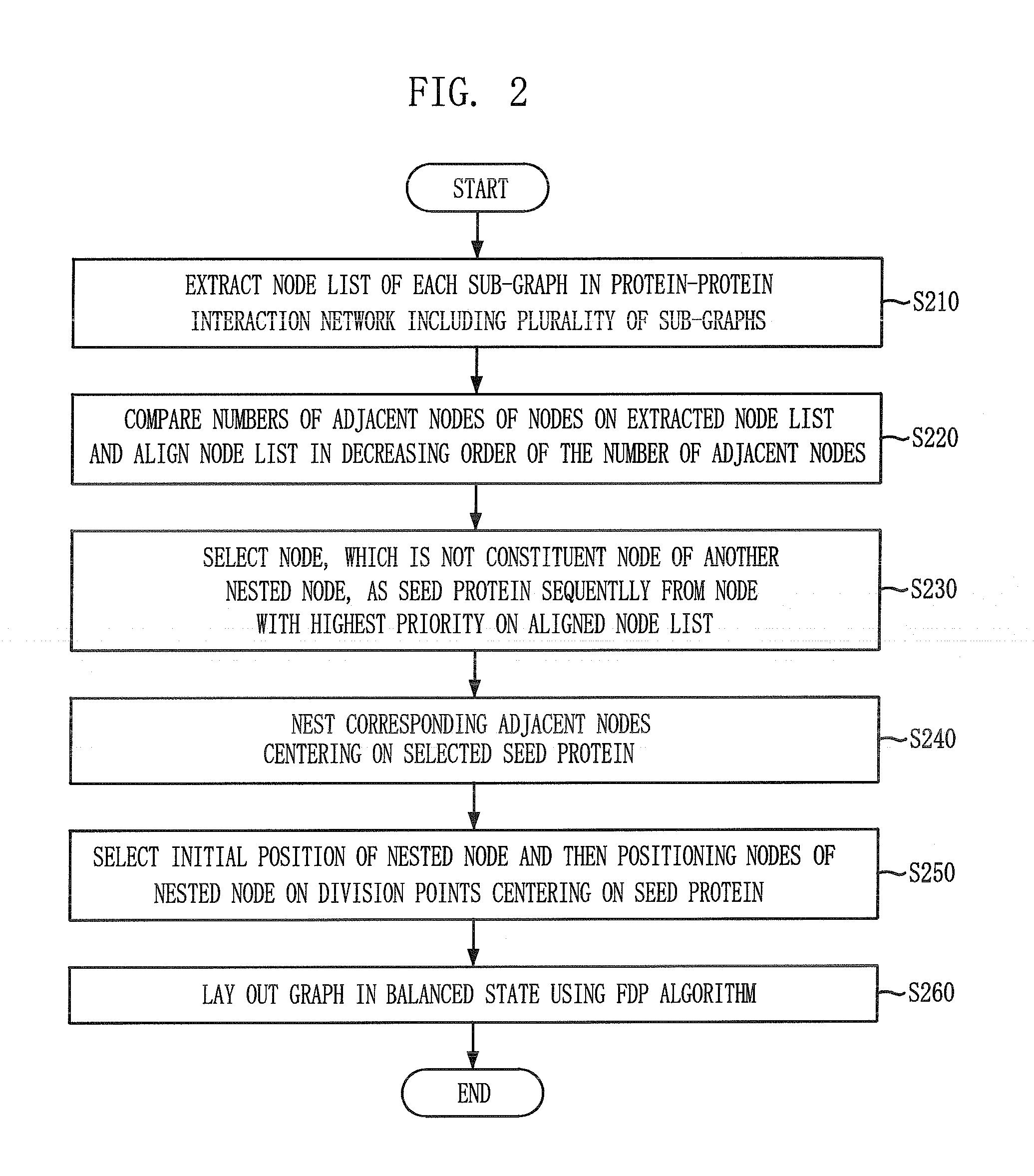
Layout method for protein-protein interaction networks based on seed protein
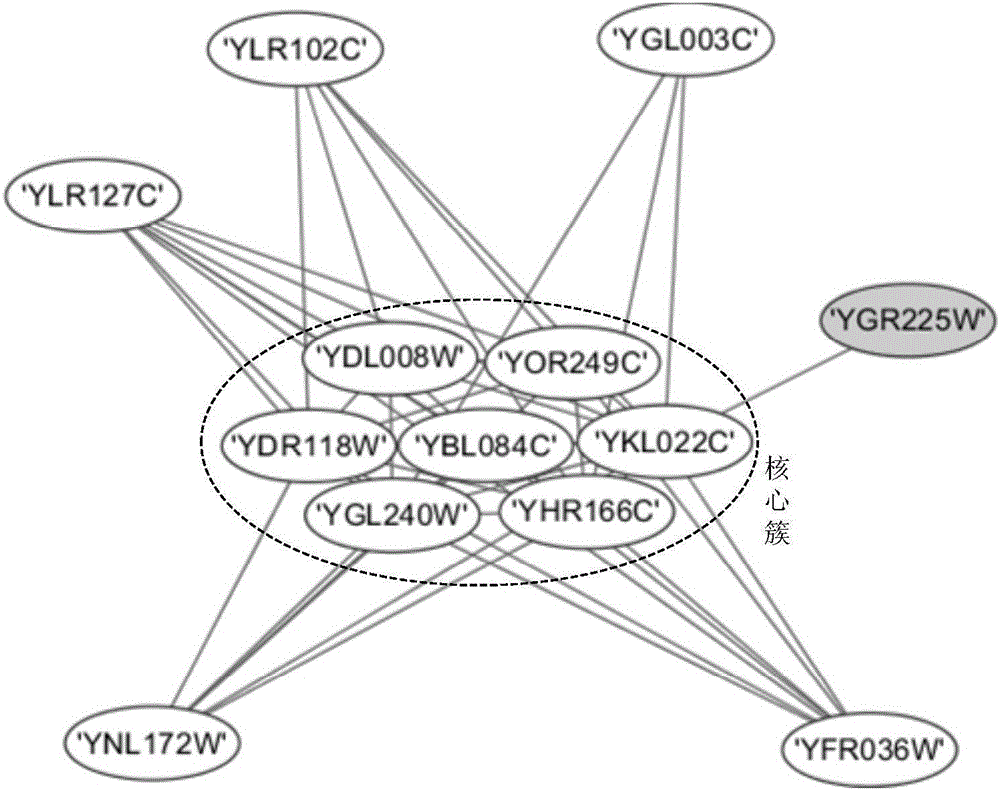
InactiveUS20080133197A1Increase speedLayout balanceData visualisationAnalogue computers for chemical processesNODALAlgorithm
Provided is a layout method for protein-protein interaction networks based on a seed protein, which is for performing multiple stages of nesting centered on a node having a high degree of physical relationship, and performing multiple stages of extension and force directed placement (FDP) with respect to a final nest graph. The layout method includes the steps of: a) extracting a node list of each sub-graph constituting a protein-protein interaction network, and aligning the node list according to adjacency of nodes; b) selecting a seed protein from the aligned node list according to node priority and nest relationship with another node; c) nesting adjacent nodes centered on the selected seed protein to generate a nested node; and d) selecting an initial position of the generated nested node, placing the nodes of the nested nodes on respective division points, centered on the seed protein, and then performing layout.
Owner:ELECTRONICS & TELECOMM RES INST
Method for identifying key protein through fruit fly optimization algorithm
ActiveCN108229643AEmbodies biological significanceImprove recognition efficiencyArtificial lifeUndirected graphProtein protein interaction network
The invention discloses a method for identifying key protein through a fruit fly optimization algorithm. The method includes the steps that a protein-protein interaction network is converted into an undirected graph, a dynamic protein-protein interaction network is constructed, the edges and nodes of the dynamic protein-protein interaction network are preprocessed, the position of a fruit fly group is randomly initialized, the food random direction and distance are searched for with the sense of smell, the taste concentration judgment value of each fruit fly individual is calculated, the odorconcentration value of each fruit fly individual is calculated, the highest odor concentration value in the current group is worked out, the fruit flies fly to the food with the sense of sight, and key protein is generated. By means of the method, the key protein can be accurately identified; the simulation experiment result shows that the method has good performance indexes including sensitivity,specificity, positive predictive value, negative predictive value, accuracy rate and recall rate harmonic value, precise value and the like; compared with other methods for identifying key protein, the method adopting the fruit fly optimization algorithm for identifying the key protein has certain advantages.
Owner:SHAANXI NORMAL UNIV
Method for identifying protein complex based on BSO (Brain Storm Optimization)
InactiveCN105590039ASignificant bioaccumulationStrong global optimization abilitySpecial data processing applicationsNODALProtein protein interaction network
The invention provides a method for identifying a protein complex based on BSO (Brain Storm Optimization); the method comprises the following steps: by utilizing strong global optimization searching capability of a BSO algorithm, regarding a protein-protein interaction network as a full network connected graph, combining gene ontology annotation function information of the protein with a topological structure of the protein-protein interaction network to define a distance among protein nodes, and carrying out preliminary clustering according to an improved k-means algorithm; then, according to four optimization searching principles of the BSO algorithm, generating a new fitness value, respectively carrying out module internal and module external optimization searching operations on a protein module which is formed preliminarily, iterating in a circulative manner and searching a most optimal global solution; and at last, carrying out post processing process. The method disclosed by the invention can keep the diversity of a group in the optimization searching process, thereby avoiding getting into local optimization; the global optimization module division is obtained, and the protein complex with remarkable biological enrichment is obtained.
Owner:HUAZHONG NORMAL UNIV
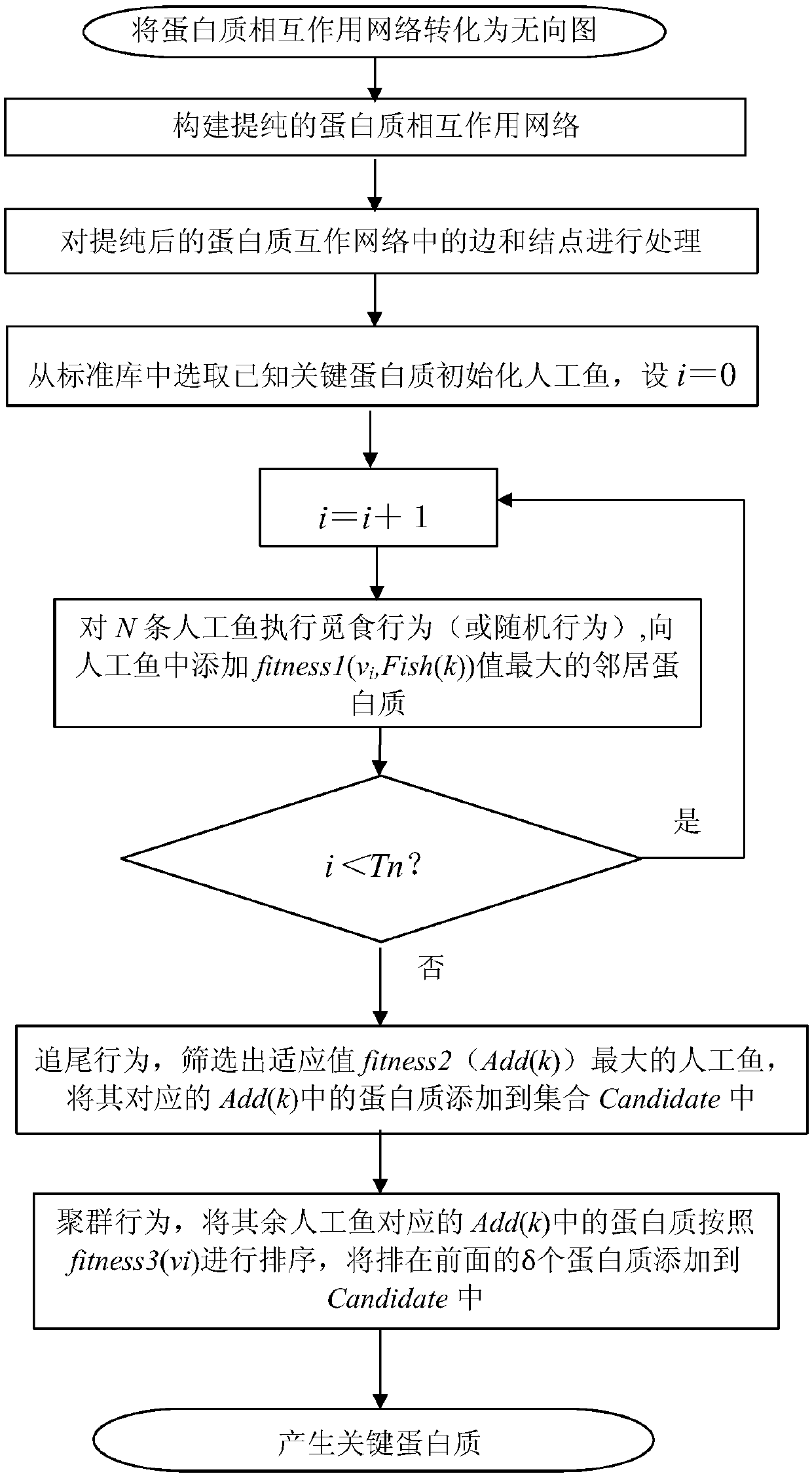
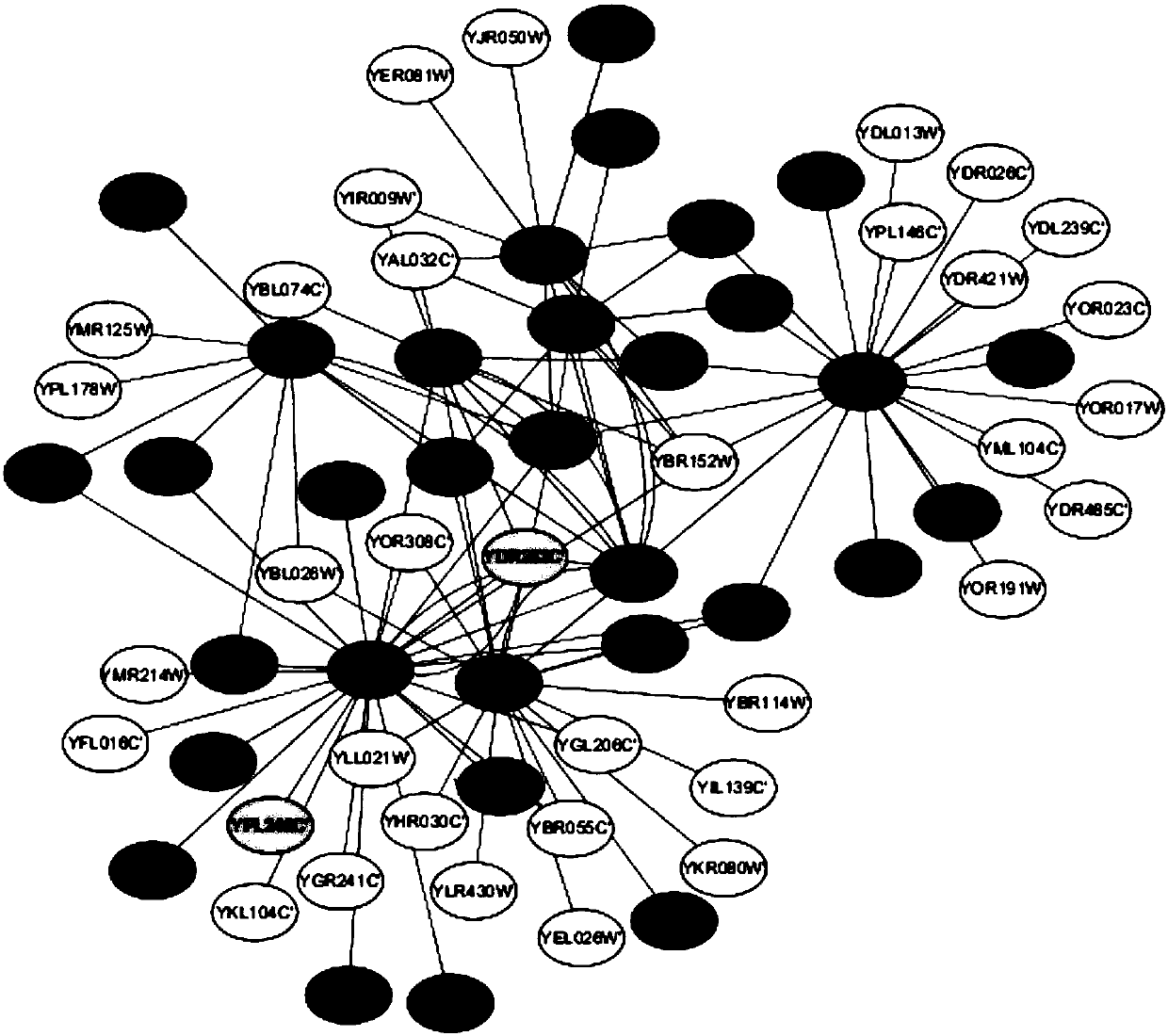
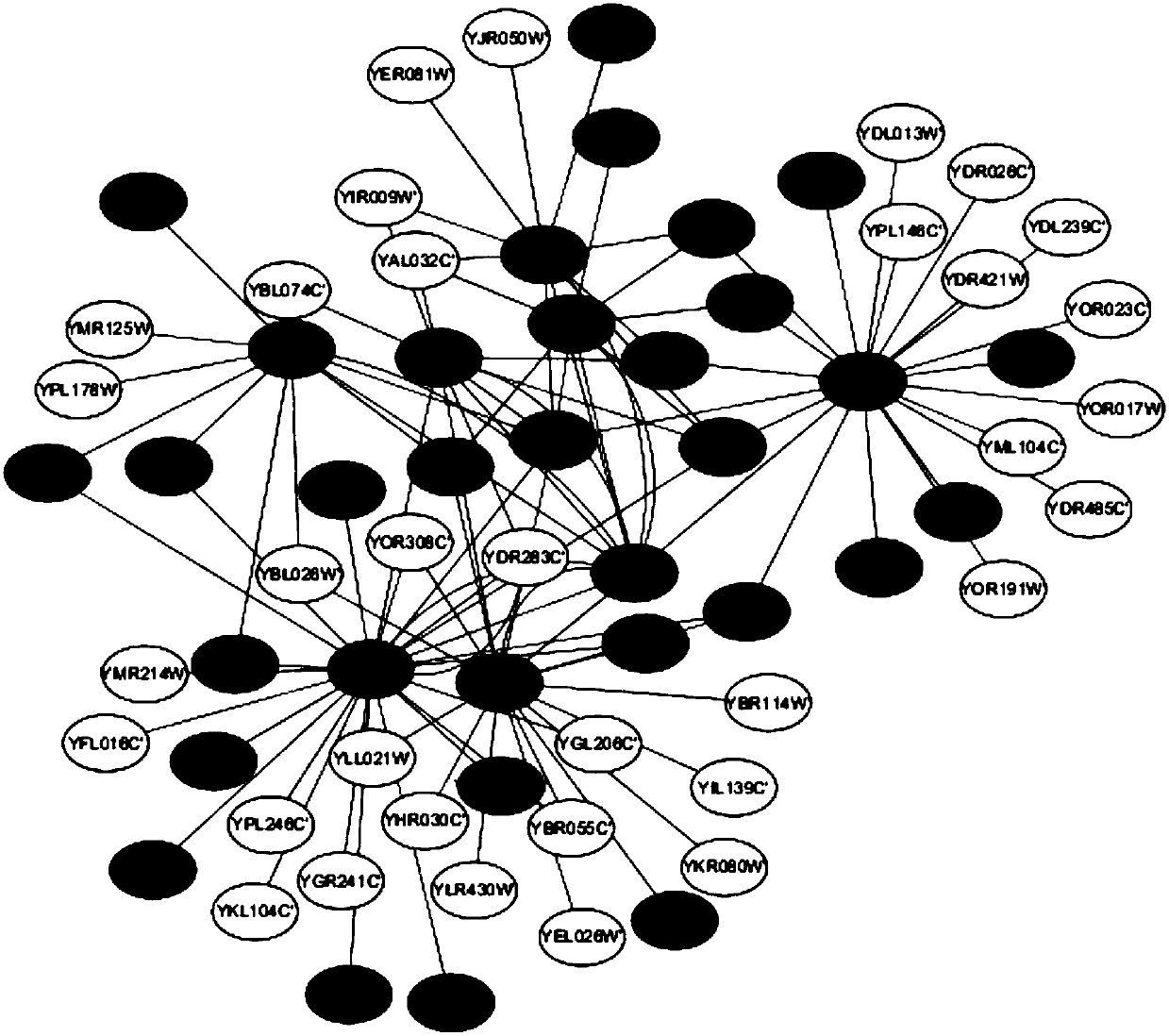
Method for identifying key proteins with AFSO (artificial fish school optimization) algorithm
ActiveCN107784196AFully consider the topological characteristicsAccurate identificationArtificial lifeProteomicsUndirected graphProtein protein interaction network
The invention discloses a method for identifying key proteins with an AFSO (artificial fish school optimization) algorithm. The method comprises steps as follows: a protein-protein interaction networkis converted into an undirected graph, a purified protein-protein interaction network is constructed, RNA gene expression values corresponding to proteins, GO comment information and degrees of proteins in known compounds are obtained, edges and nodes of the purified protein-protein interaction network are treated, known key proteins are selected as initial artificial fishes, the artificial fishes execute foraging behavior, random behavior, following behavior and swarm behavior, and the key proteins are produced. According to the method, the key proteins can be identified accurately; a simulation experiment result indicates that performance of indexes such as sensitiveness, specificity, a positive predictive value, a negative predictive value and the like is better; compared with other methods for identifying the key proteins, the method has the advantages that optimizing characteristics of artificial fish schools are combined with topological characteristics of the protein-protein interaction network to realize the key protein identification process, and the accuracy rate of the key protein identification is increased.
Owner:SHAANXI NORMAL UNIV
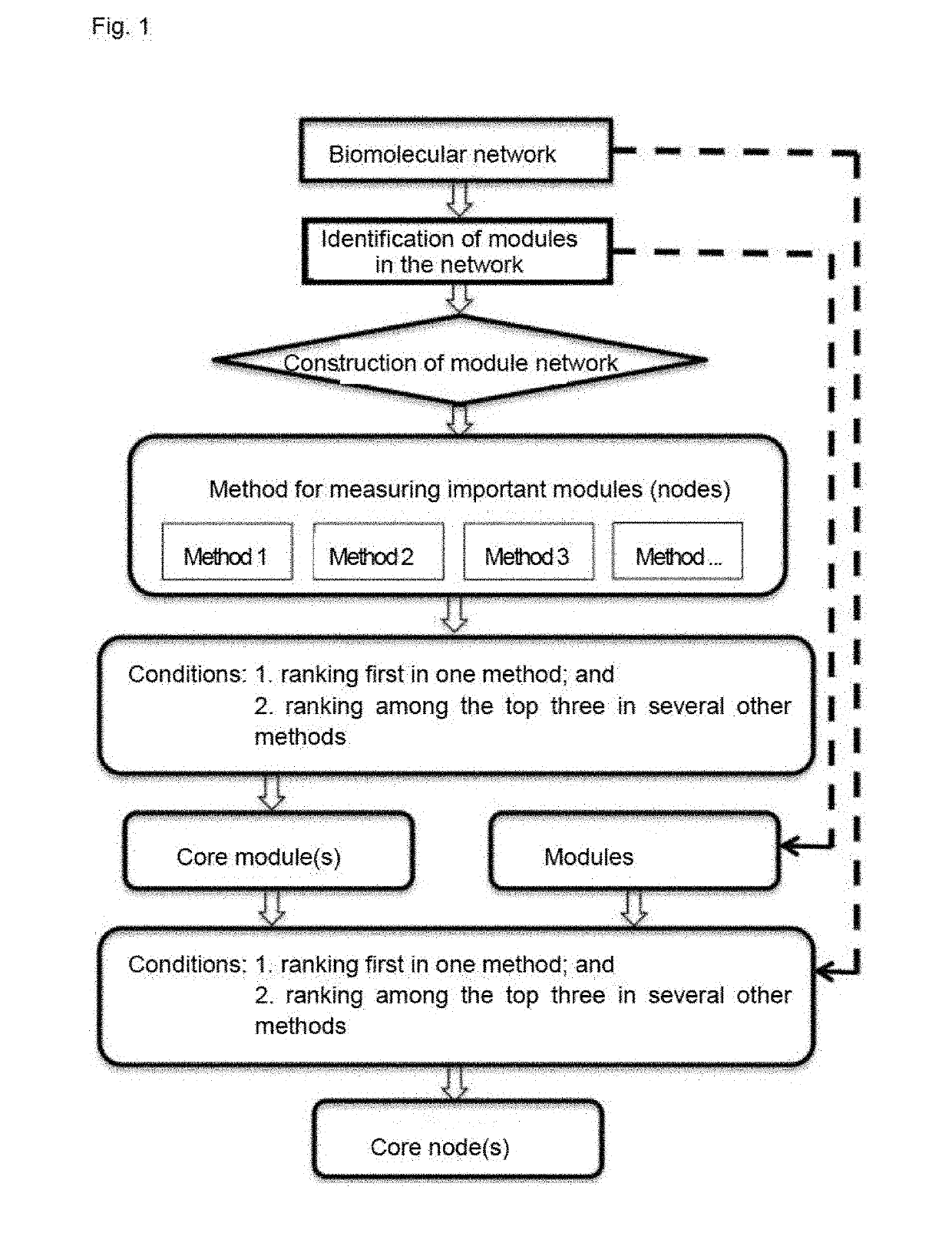
Method for identifying key module or key node in biomolecular network
PendingUS20190139621A1Strong integrationData visualisationBiostatisticsRegulation of gene expressionStructure of Management Information
A method for identifying a key module and a key node of a biomolecular complex network by fusing multiple methods on the basis of a topological structure of the biomolecular complex network which may be such as a protein-protein interaction network, a gene expression regulation network, a biological metabolism network, an epigenetic network, a phenotypic network or a signaling network, comprising the following main steps: according to the biomolecular network and a module division for the network, comprehensively and quantitatively identifying the key module and key node on the basis of the topological structure by various measuring methods from multi-angle.
Owner:WANG ZHONG +1
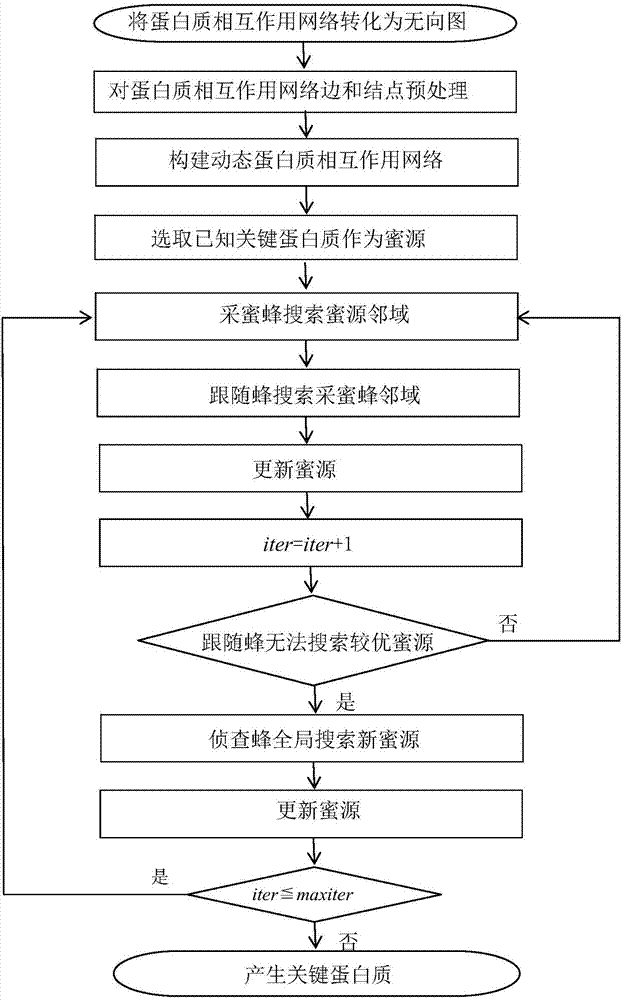
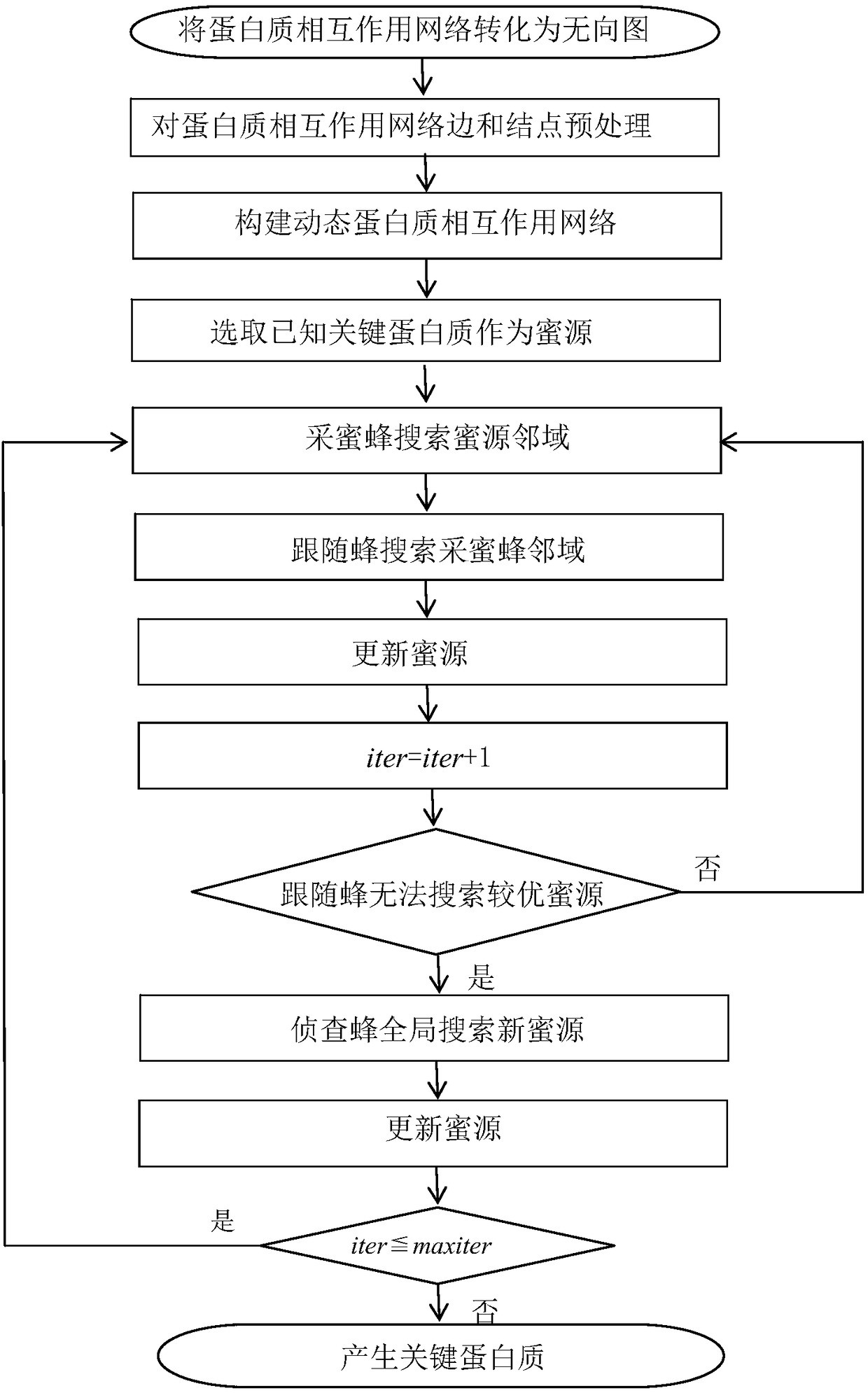
Method for identifying key protein using artificial bee colony optimization algorithm of foraging mechanism
InactiveCN106874708AFeaturesSolve the shortcomings of not being able to consider the overall nature of the networkArtificial lifeProteomicsProtein protein interaction networkPerformance index
The invention discloses a method for identifying key protein using an artificial bee colony optimization algorithm of a foraging mechanism. The method comprises: converting a protein-protein interaction network to an undirected graph, obtaining ribonucleic acid genetic expression values corresponding to protein, preprocessing edges and nodes of the protein-protein interaction network, establishing a dynamic protein-protein interaction network, selecting known key protein as a honey source, honey bees searching neighbourhood of the honey source, following bees searching neighbourhood of the honey bees, updating the honey sources, investigating bees searching new honey sources in a global manner, updating the honey sources, and generating the key protein. The method can accurately identify the key protein. Results of simulation experiments show that sensitiveness, specificity, positive predictive values, negative predictive values and other performance indexes are relatively excellent. Compared with other key protein identification method, the identification process of key protein realized by combining optimizing characteristics of artificial bee colony with characteristics of the protein-protein interaction network improves identification accuracy rate of the key protein.
Owner:SHAANXI NORMAL UNIV
Method for establishing protein-protein interaction network by utilizing text data
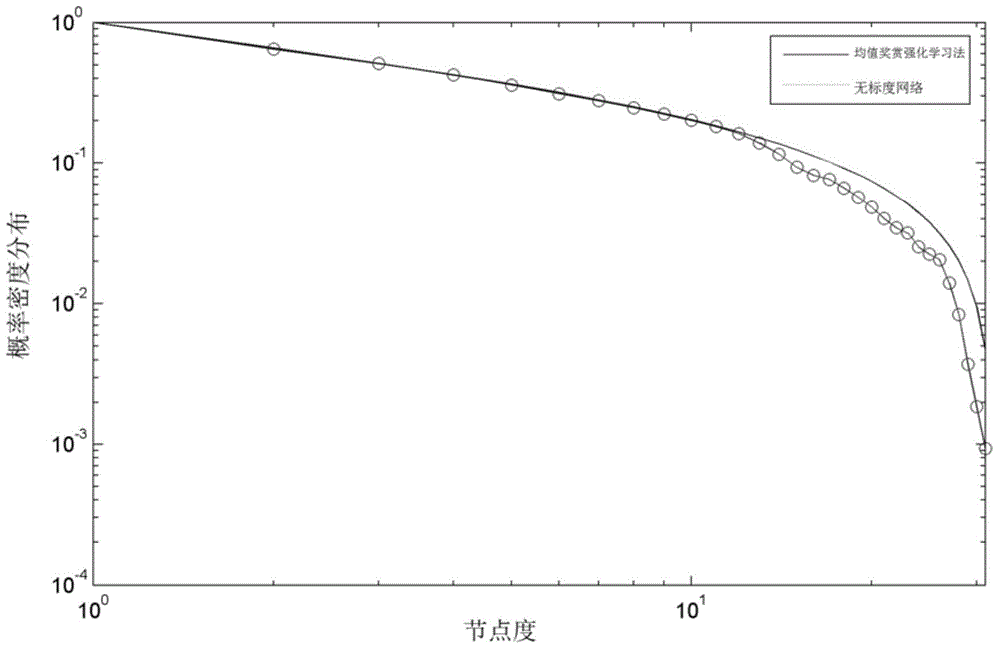
InactiveCN104657626AEnsuring scale-free propertiesIncrease or decrease boundary valueProteomicsGenomicsNegative feedbackProtein protein interaction network
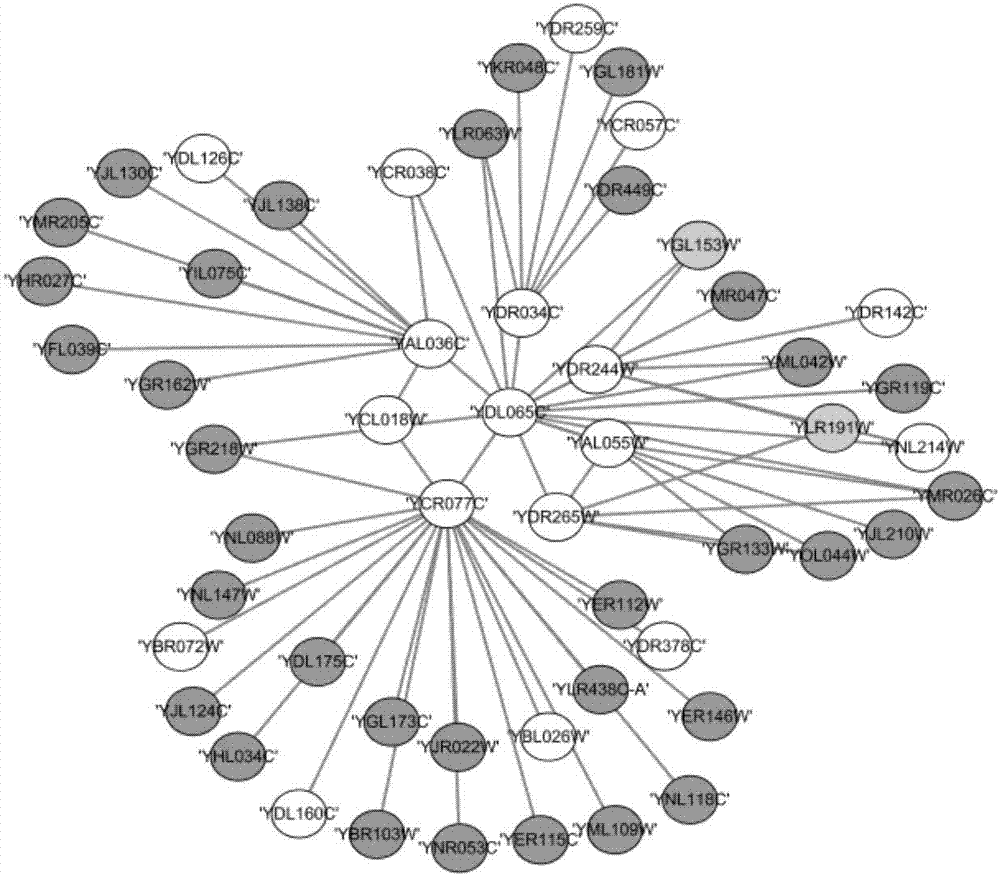
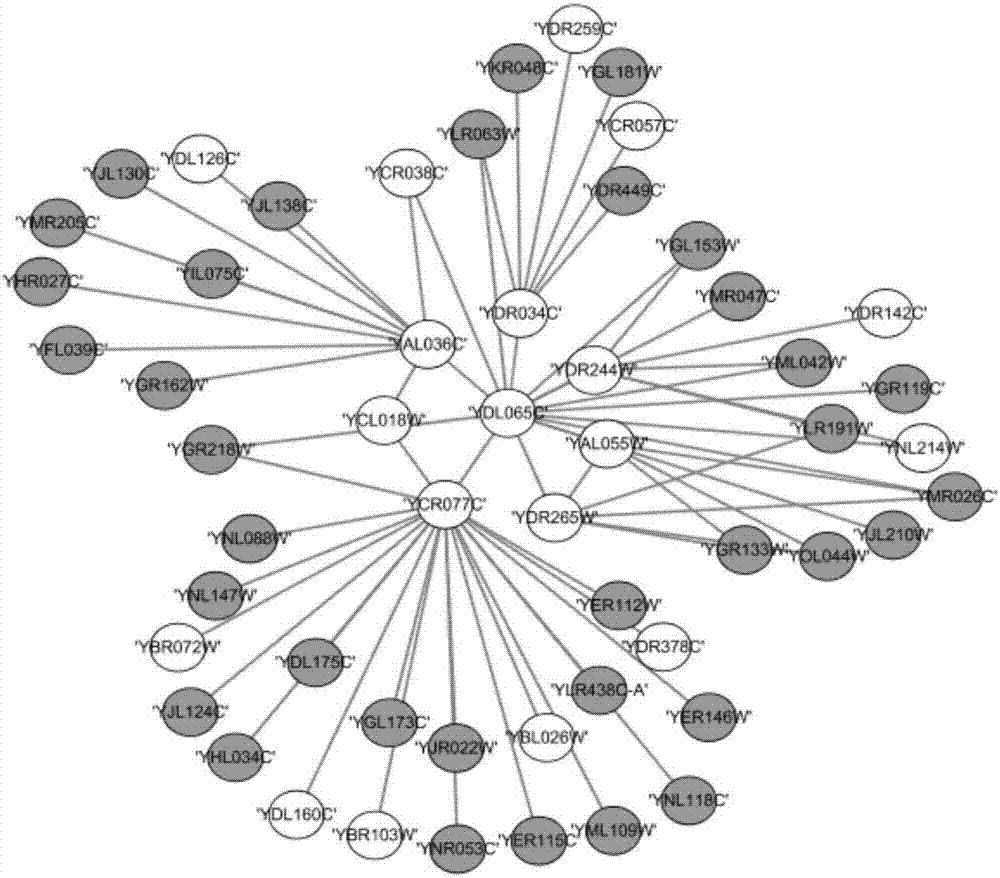
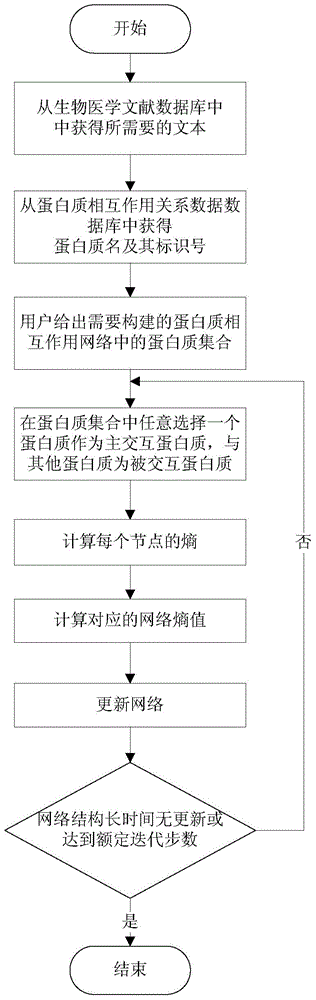
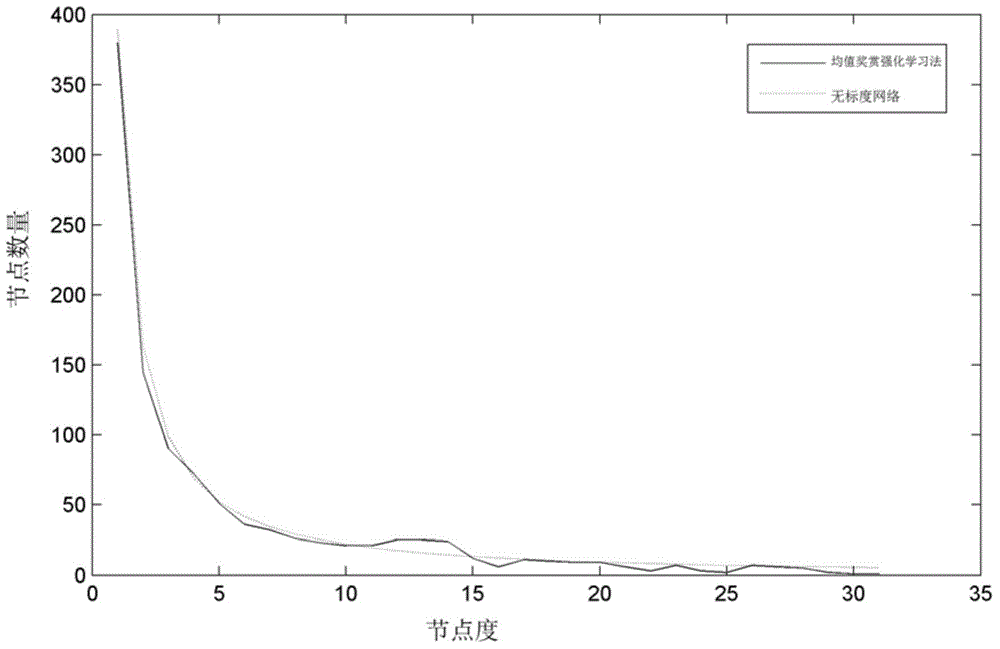
The invention discloses a method for establishing a protein-protein interaction network by utilizing text data. The method is characterized by comprising the following steps: (1) establishing a protein set; (2) recording the probability values of interactions of every two proteins in the protein set; (3) establishing an initial network structure according to the probability values; (4) repeatedly selecting protein, giving feedback values of positive or negative reactions, iterating discontinuously on the initial network structure to obtain a final network structure. According to the method, the probability graph of a reaction network is established through reinforcement learning in the mode of repeatedly selecting and interacting on the basis of positive feedback, negative feedback and prohibited feedback; the probability graph is combined seamlessly with biological knowledge and biological data.
Owner:SUZHOU UNIV
A method and system for determining cancer network markers based on probability model
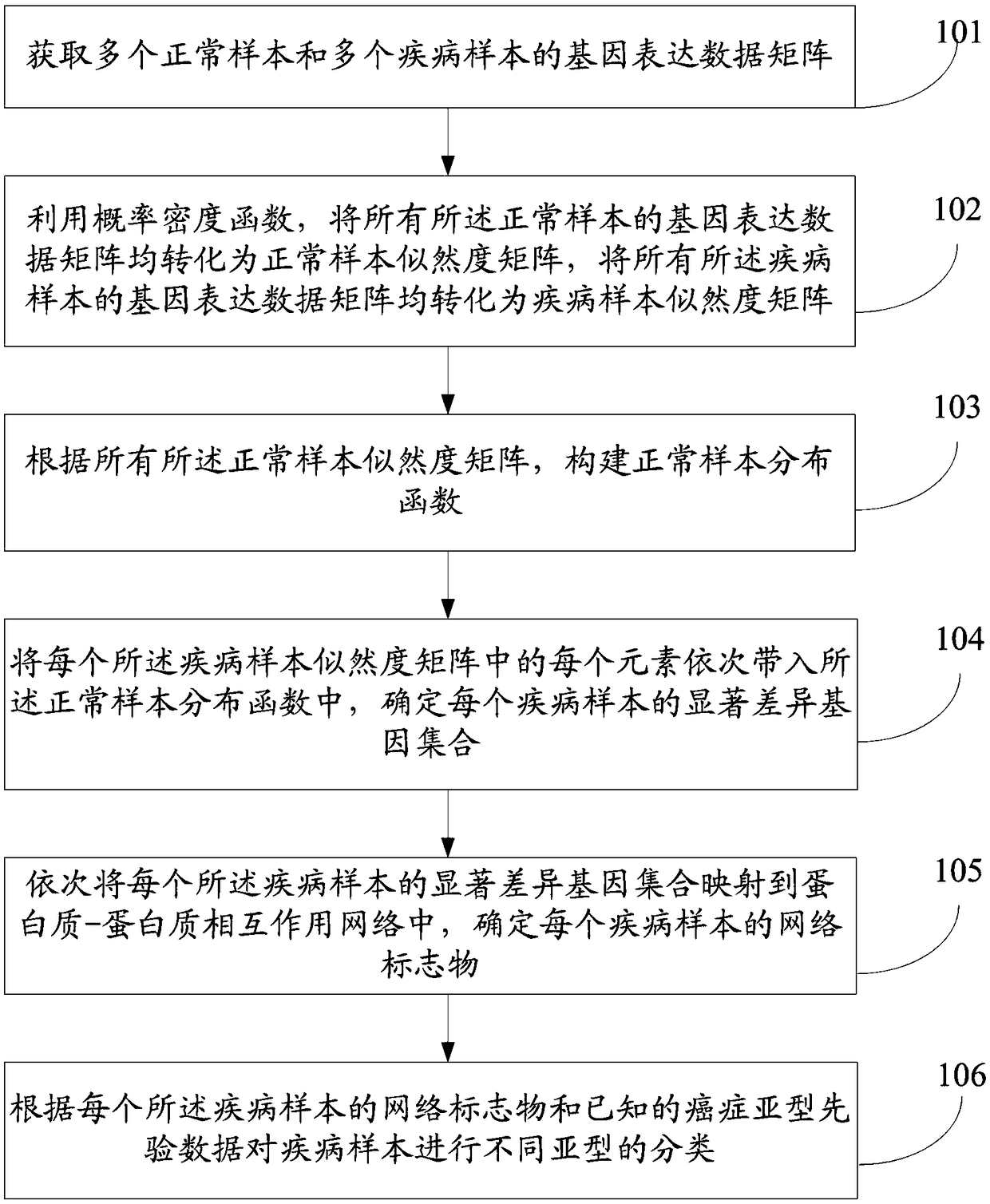
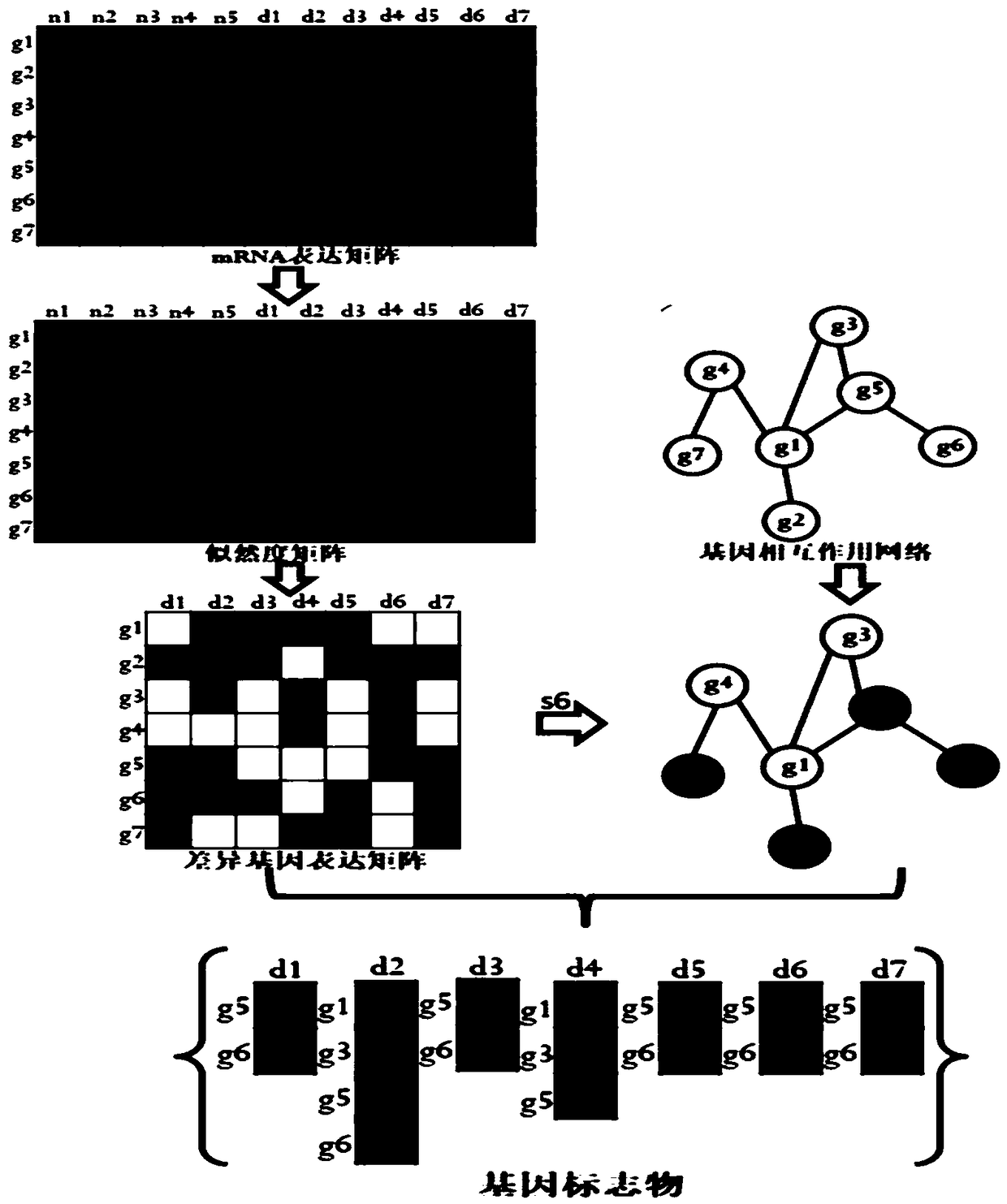
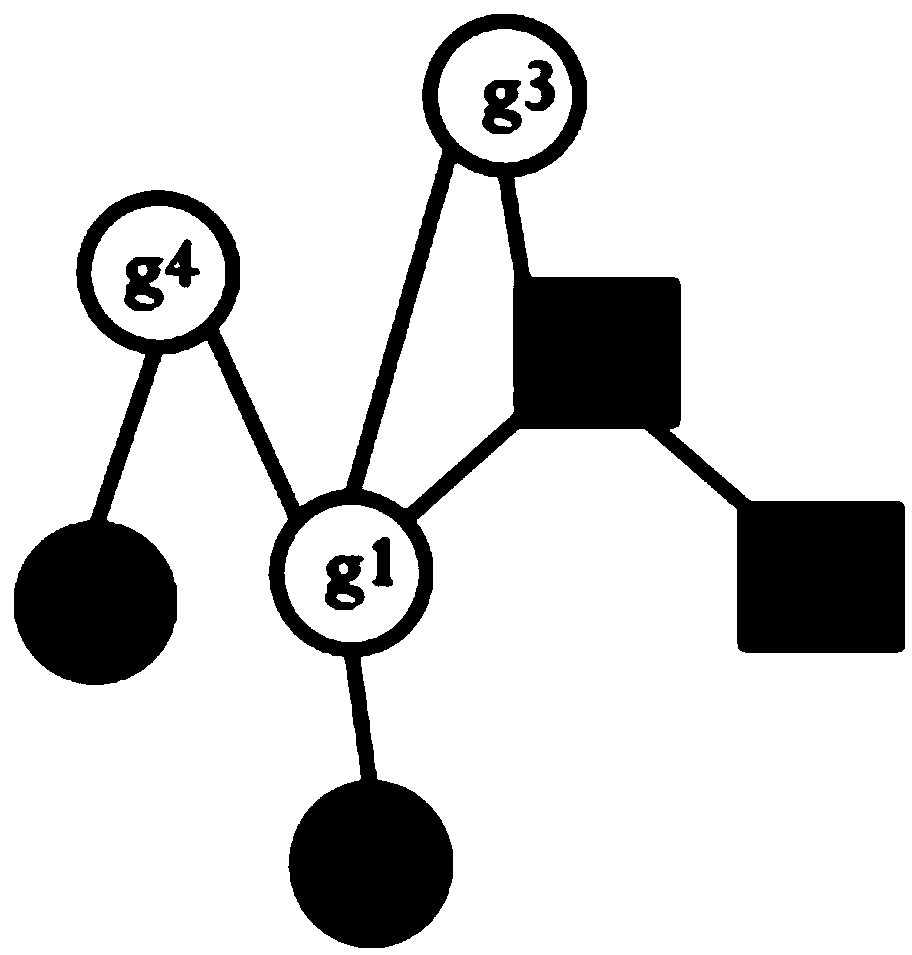
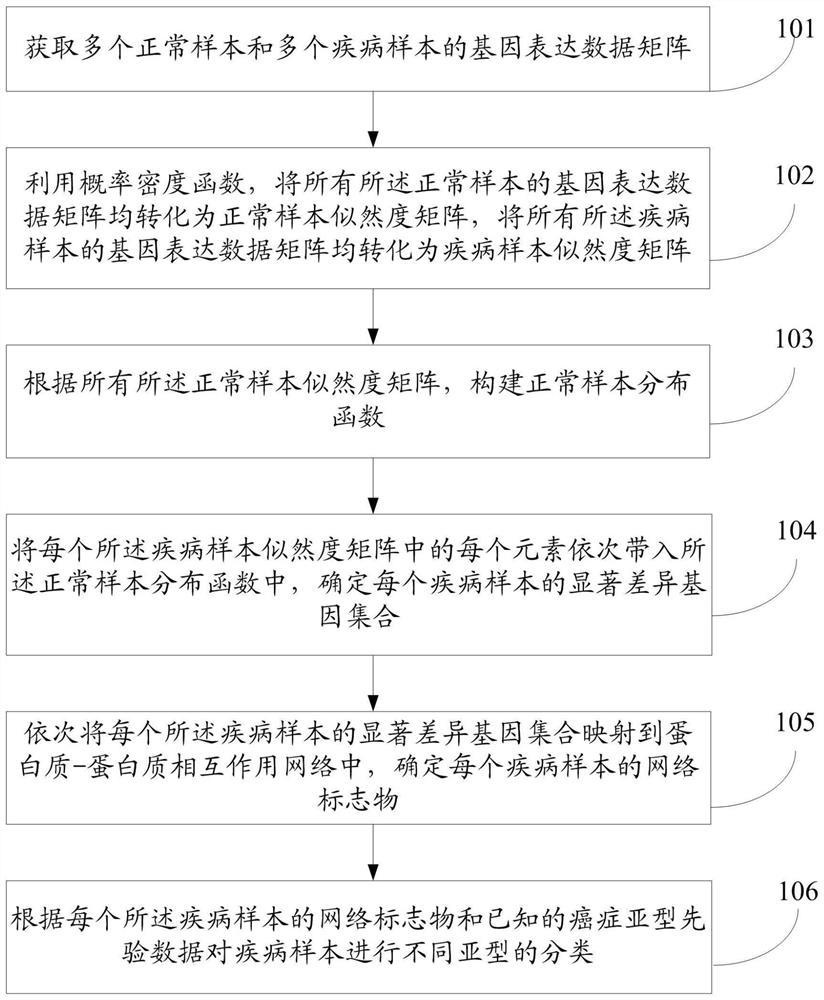
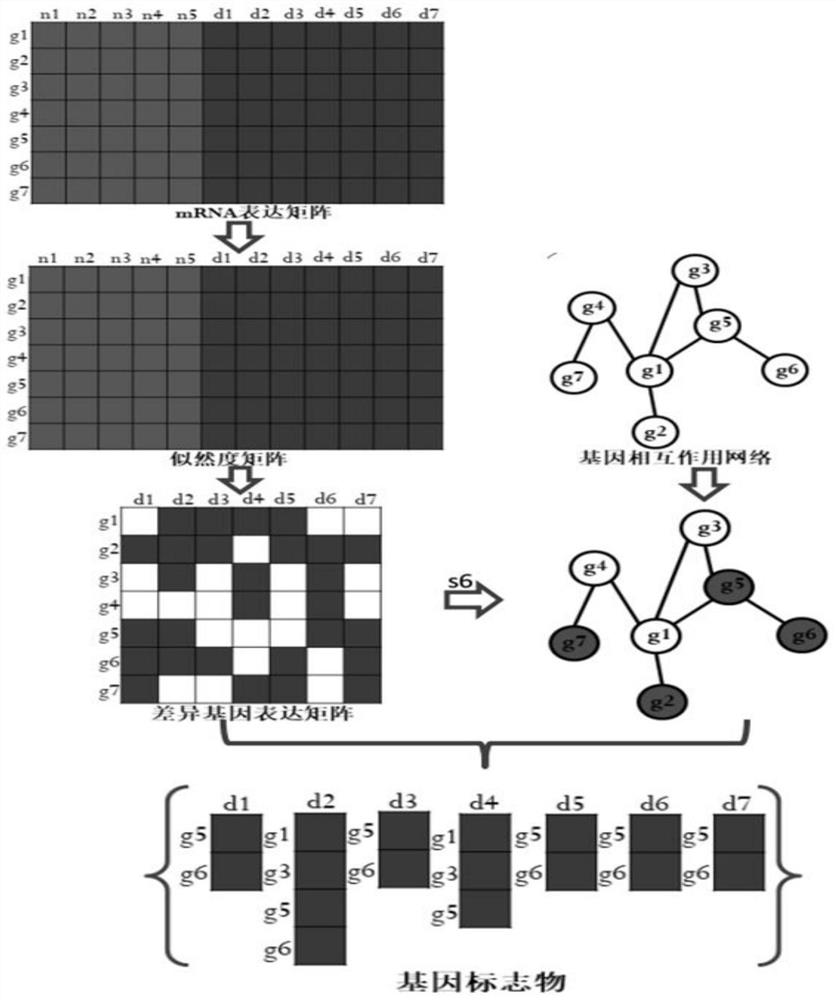
InactiveCN109101783AGet accurate and effectiveAccurate diagnosisSpecial data processing applicationsDiseaseNormal density
The invention discloses a method and a system for determining cancer network markers based on a probability model. The method comprises the following steps: utilizing a probability density function, converting gene expression data matrices of all normal samples and disease samples obtained into likelihood matrices, and constructing a normal sample distribution function according to the likelihoodmatrices of all normal samples; each element in the likelihood matrix of each disease sample is then brought into the normal sample distribution function, the set of significantly different genes foreach disease sample is determined, and the set of significantly different genes for each disease sample is mapped to proteins. Protein-protein interaction networks, and network markers for each disease sample are identified. The method or system provided by the invention can accurately and effectively acquire cancer network markers, and utilize the cancer network markers to carry out subtype classification of diseases so as to realize precise diagnosis and treatment of diseases.
Owner:WENZHOU UNIVERSITY
Method for predicting function module and function on basis of PPI network hierarchy structure
ActiveCN107798215AImprove reliabilityFunction increaseProteomicsGenomicsAlgorithmProtein protein interaction network
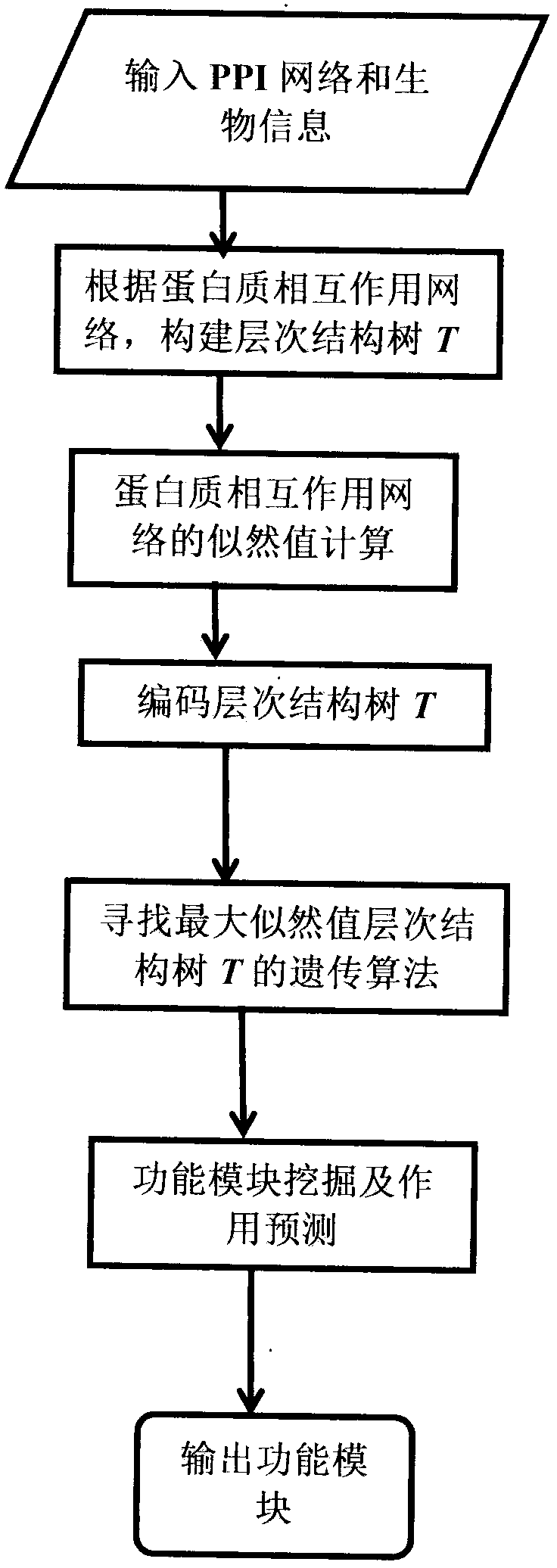
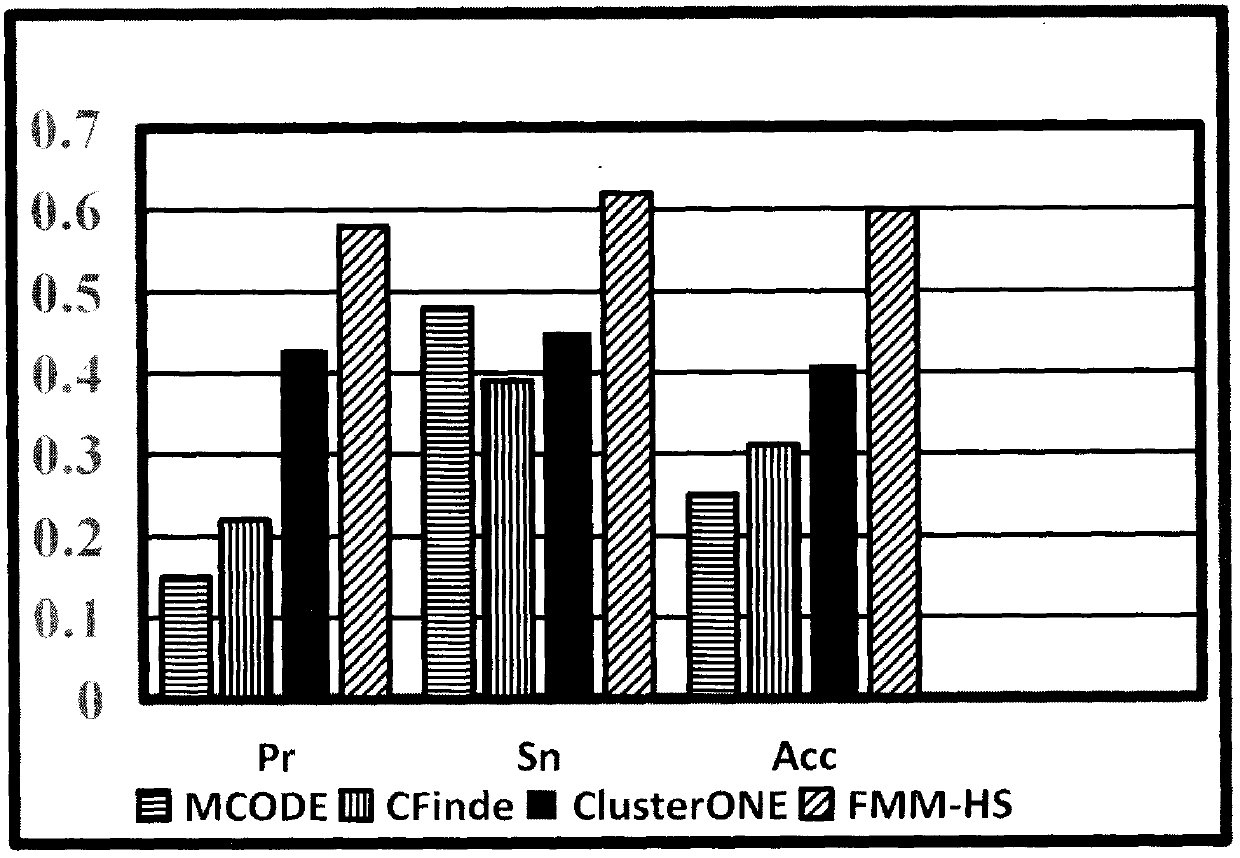
The invention relates to a method for predicting a function module and function on the basis of the PPI network hierarchy structure. According to the technical scheme, the method comprises the steps that PPI network and biological information are input, a hierarchy tree T is built according to a protein-protein interaction network, the hierarchy tree T is coded according to likelihood value calculation of the protein-protein interaction network, a genetic algorithm of a maximum likelihood value hierarchy tree T is sought, and function module mining and function prediction are conducted. Accordingly, the defects that in the sparse PPI network low in density, the effect is poor, and randomness exists are overcome. According to the method, mining and function prediction are conducted on the function module according to the maximum likelihood value hierarchy tree T, and function module mining and function prediction are conducted simultaneously through network likelihood value calculation;on the basis of considering network topology, corresponding biological information is blended, an internal relationship among network nodes is reflected, many unnecessary density calculations are reduced, the prediction result is more accurate, and the reliability of the prediction result is improved.
Owner:YANGZHOU UNIV
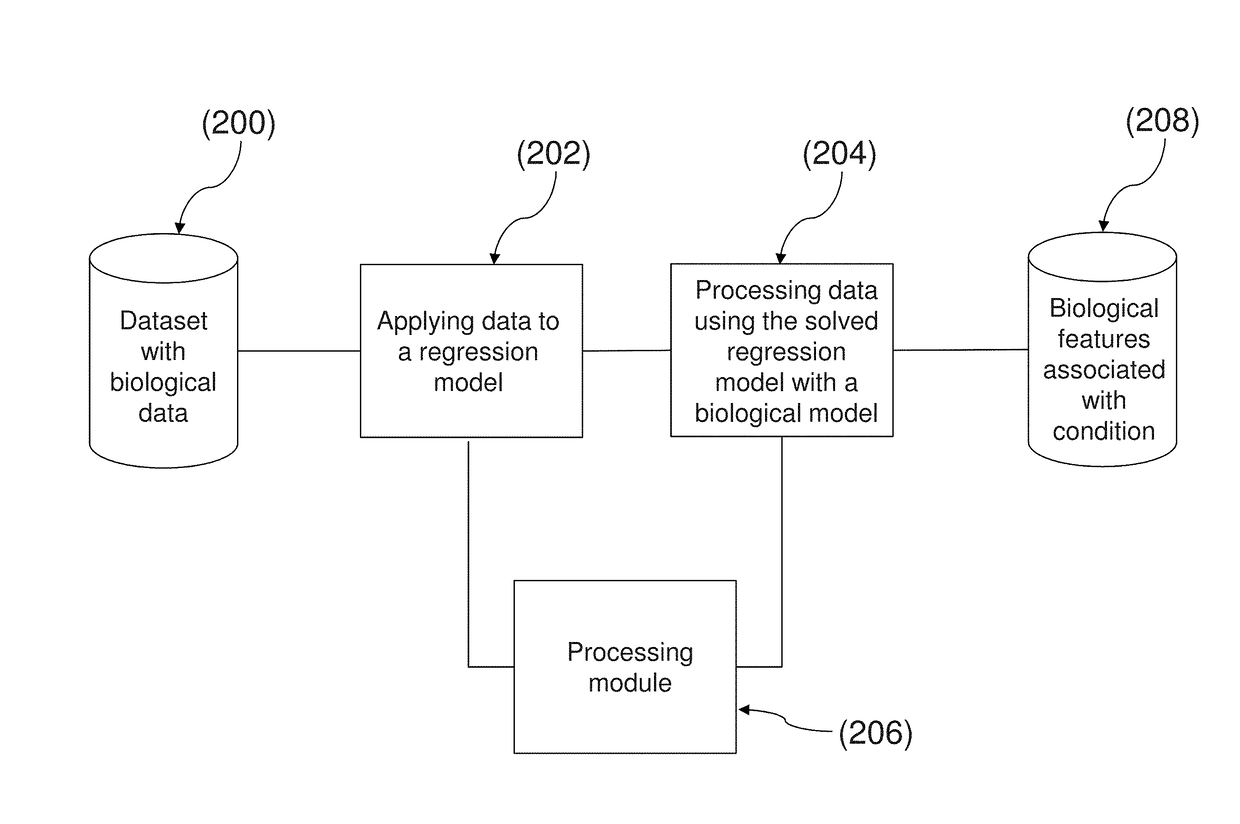
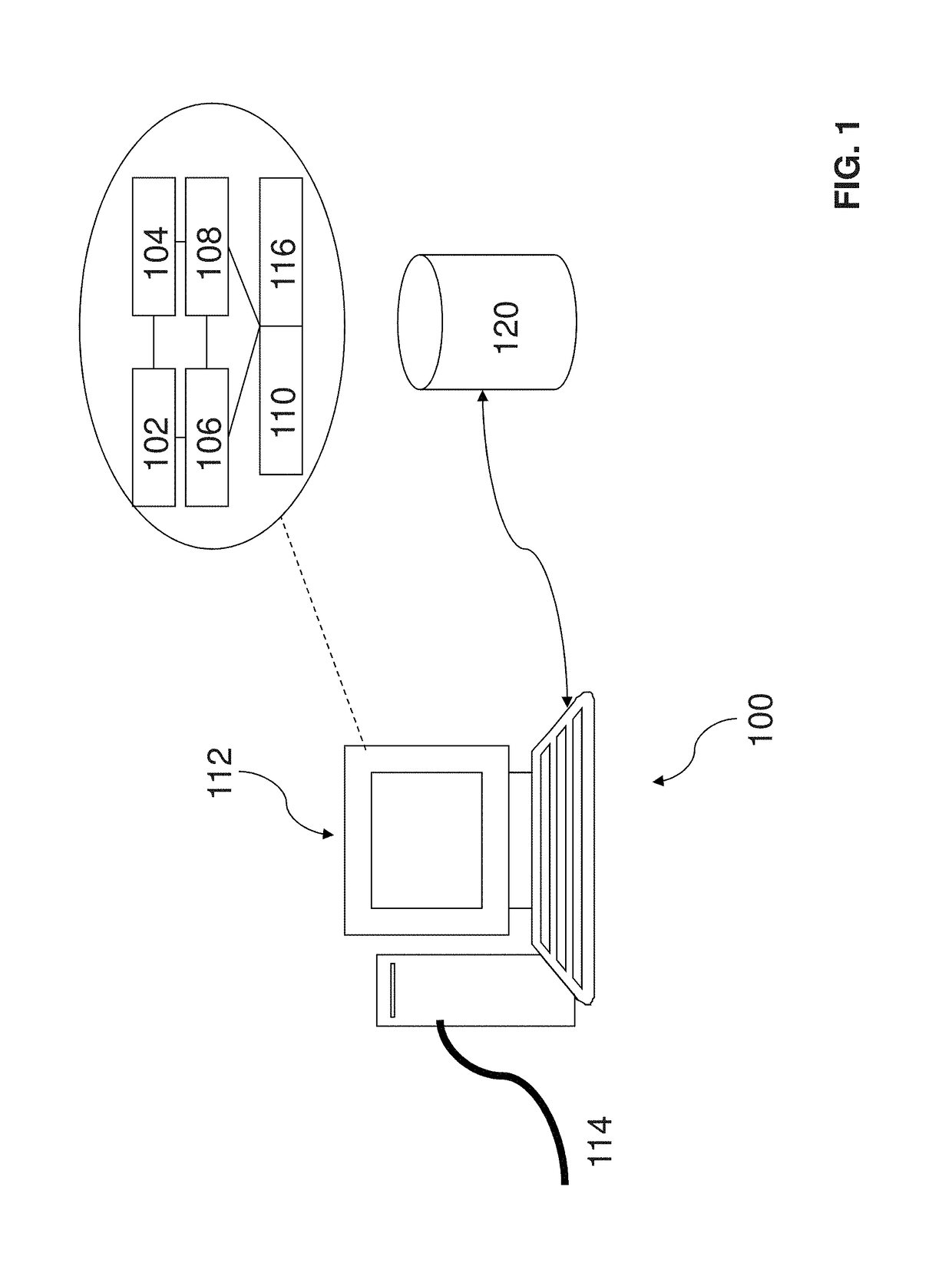
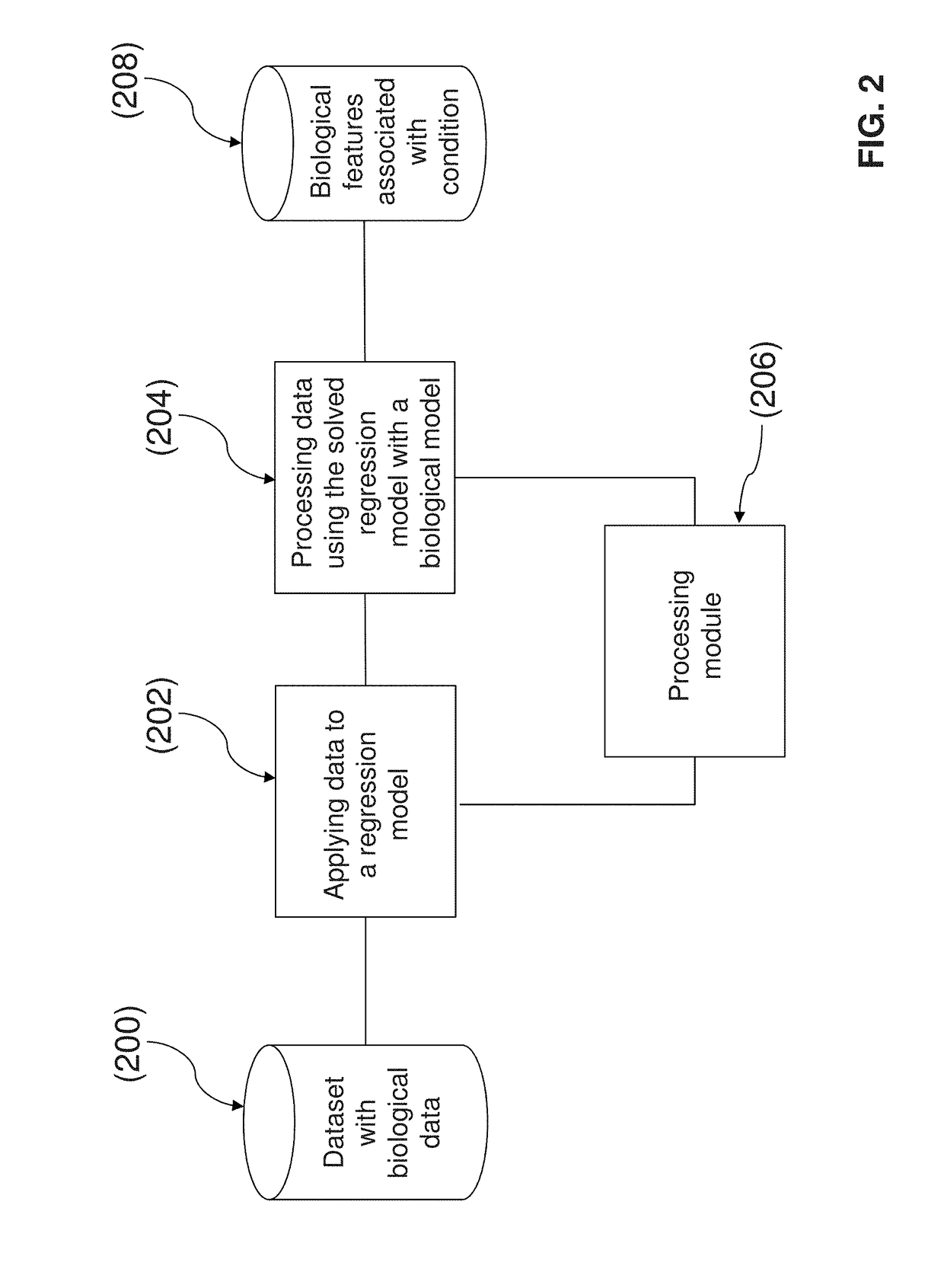
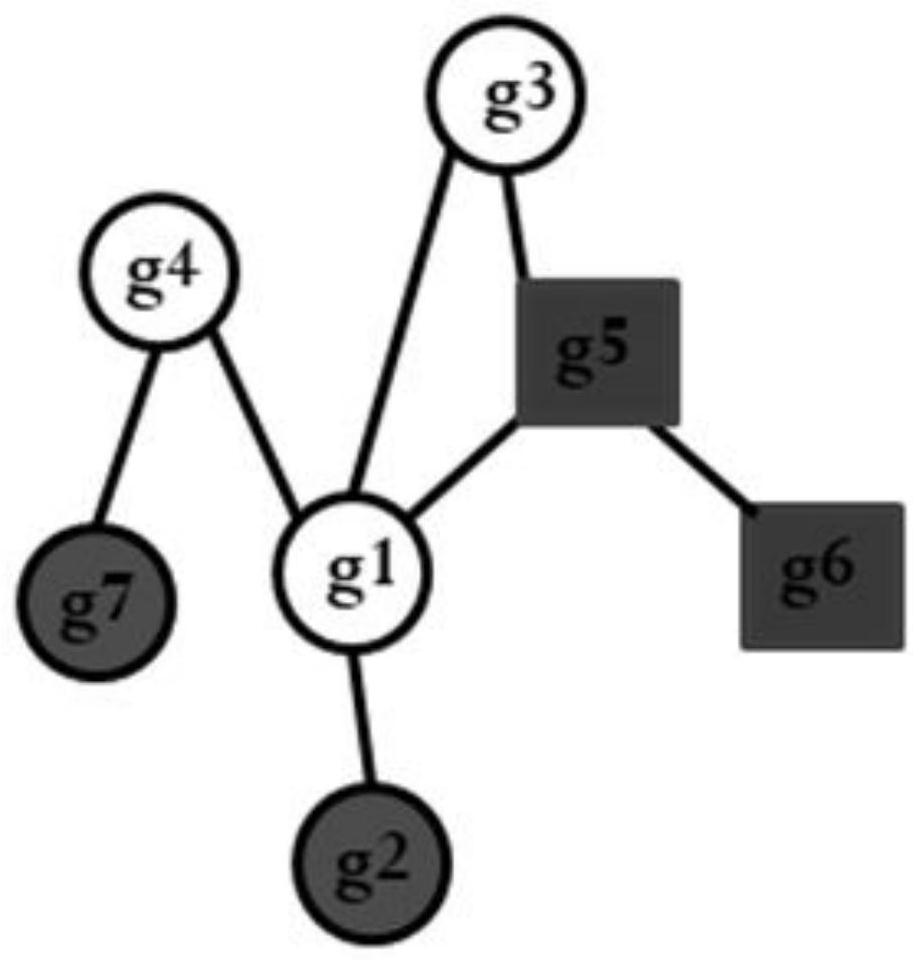
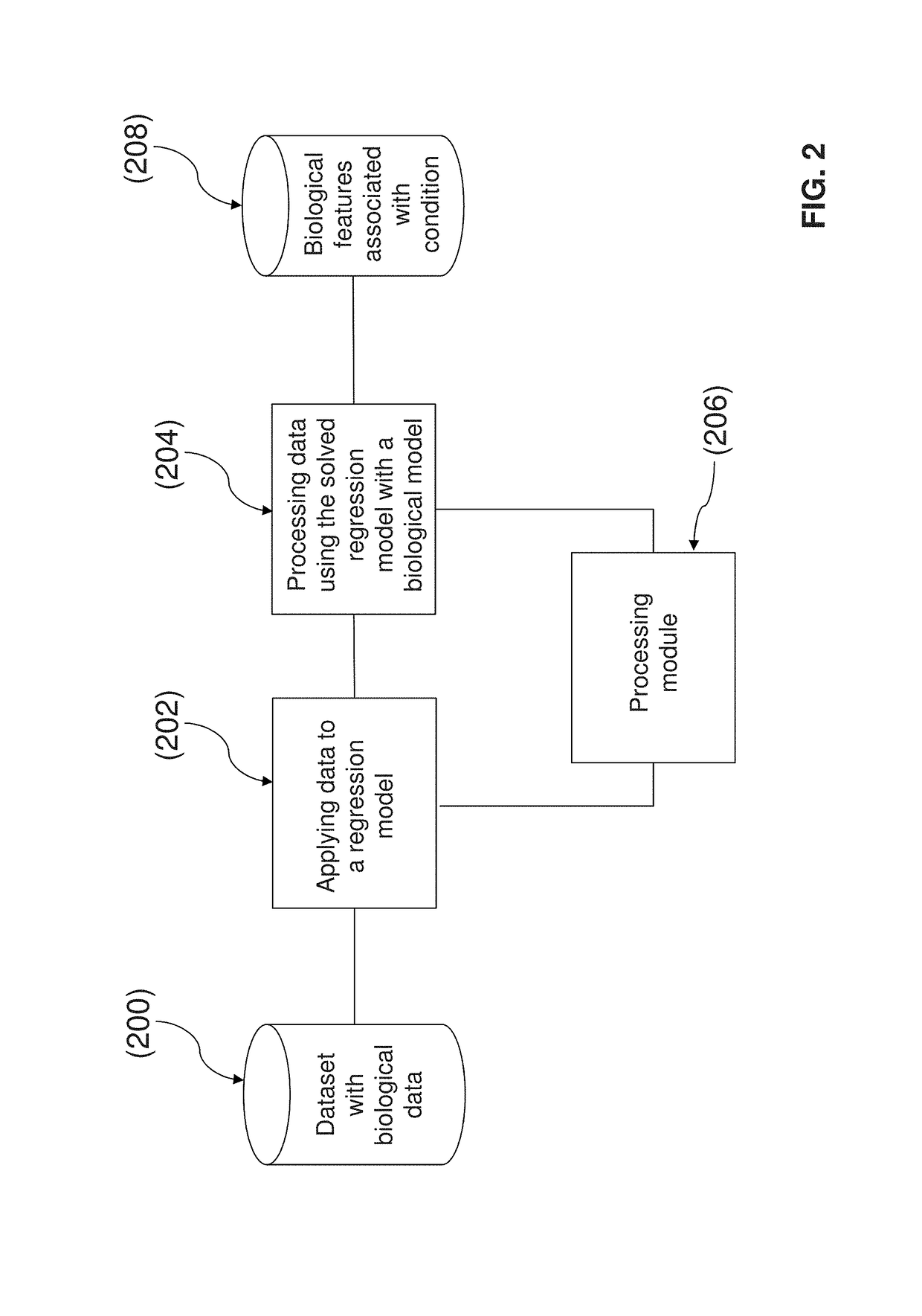
System and method for determining an association of at least one biological feature with a medical condition
A system and a method for determining an association of at least one biological feature with a medical condition, in particularly, but not exclusively, a system and a method of determining an association of at least one biological feature in form of a gene expression with cancer or a subtype of cancer that can include the generation of a simplified protein-protein interaction network based on processed biological data. The system and respective method is especially suitable for analysis of high dimensional and low sample size biological datasets such as in cancer research.
Owner:MACAU UNIV OF SCI & TECH
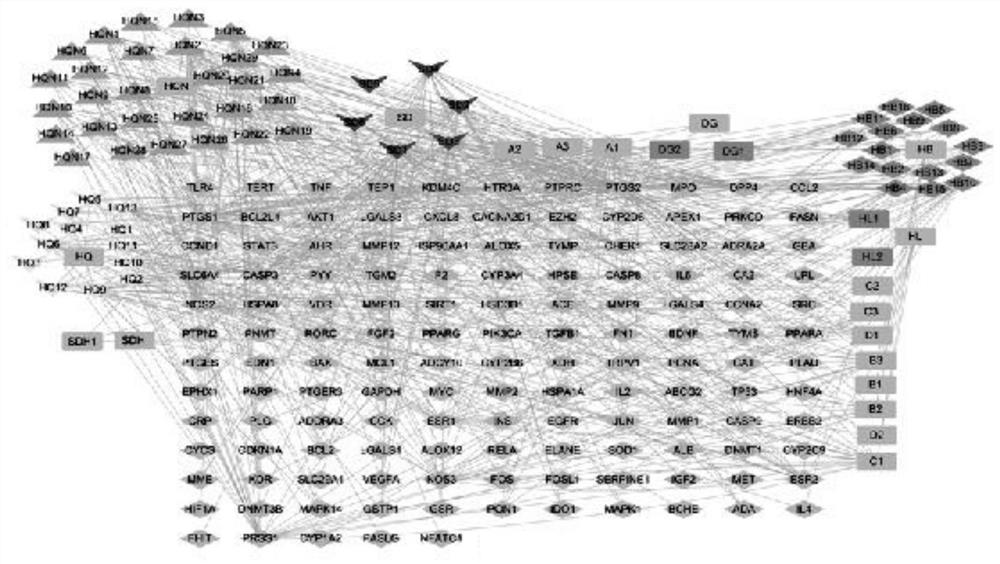
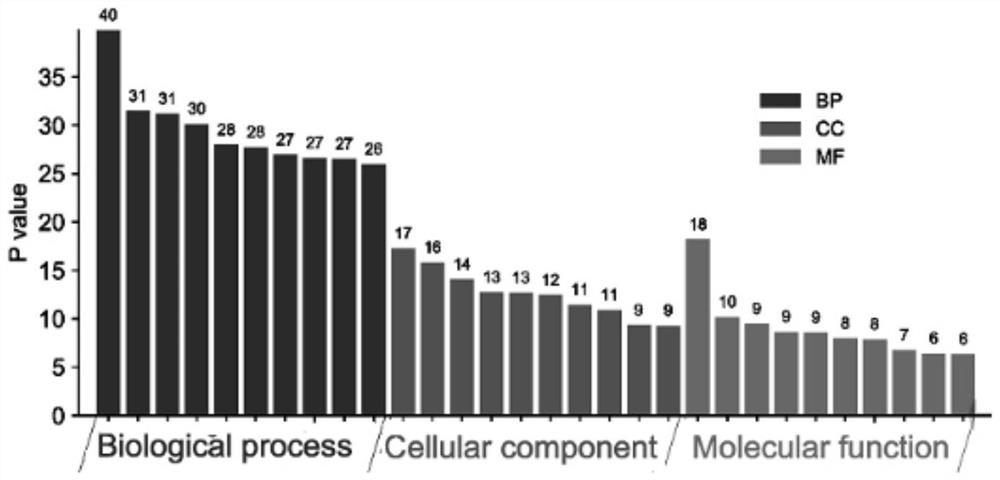
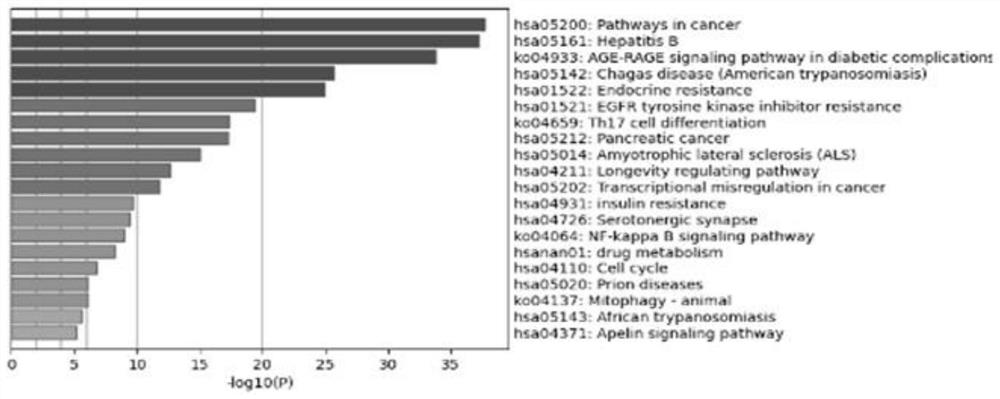
Research method of action mechanism of Chinese angelica and six-yellow decoction for treating ulcerative colitis
The invention relates to a research method of an action mechanism of Chinese angelica and six-yellow decoction for treating ulcerative colitis. The intersection target of the active component and the disease is obtained through database analysis of TCMSP, uniprot and the like. A protein-protein interaction (PPI) network is constructed based on String, and a result is visually analyzed by using Cytoscape software to obtain important modules and core targets in the PPI network. Gene GO and KEGG pathway enrichment analysis are performed on a target spot by adopting Metascape, to obtain a component-target spot-gene function-pathway-disease visual graph, and an action mechanism is analyzed. The method provides a new method for researching the drug-target interaction mechanism of the Chinese angelica and six-yellow decoction for treating ulcerative colitis; the problems that the number of interactions between multiple active ingredients and multi-target protein is too large and screening is difficult are solved, and the method has important application significance for guiding research and development of traditional Chinese medicine compounds.
Owner:SUN YAT SEN UNIV +1
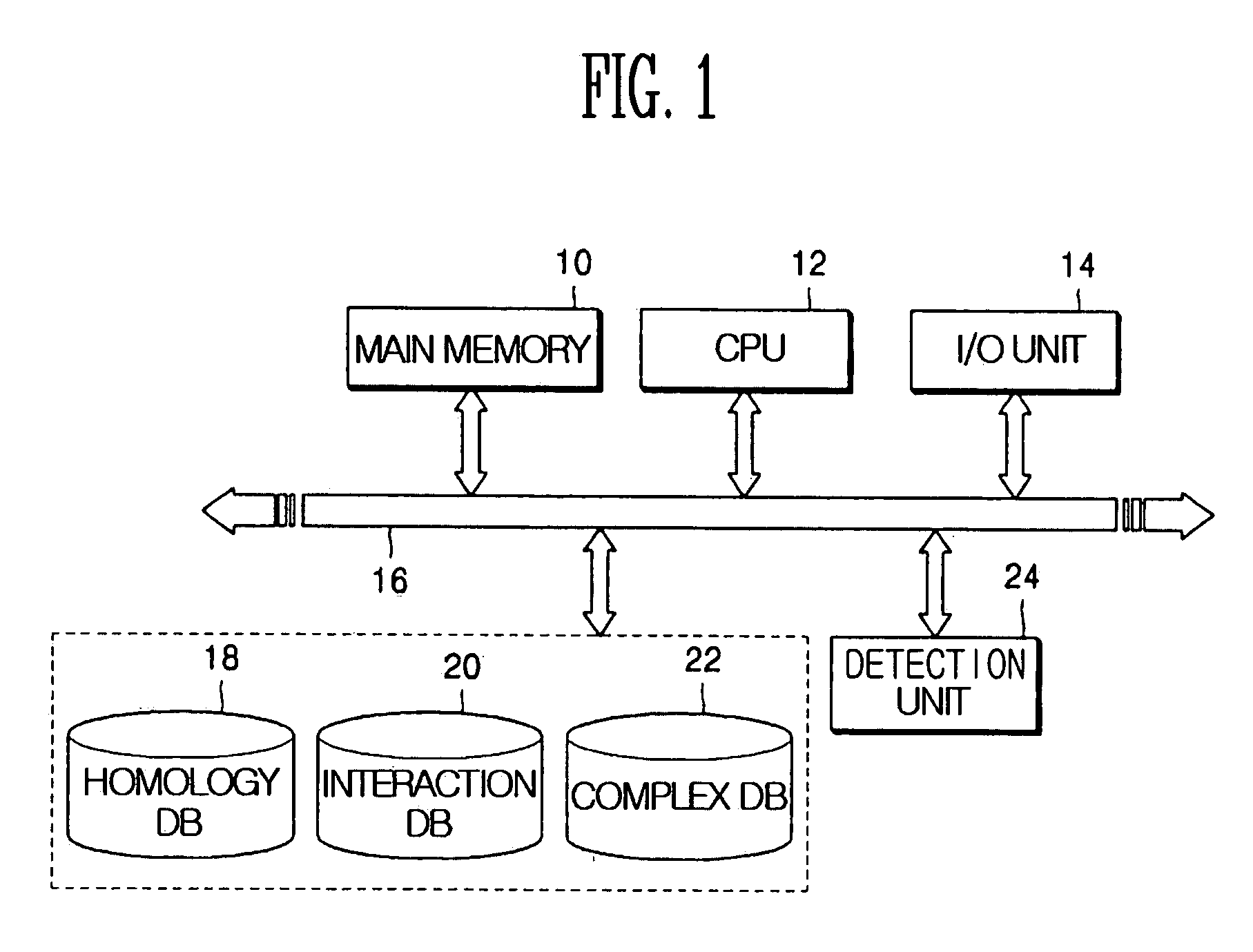
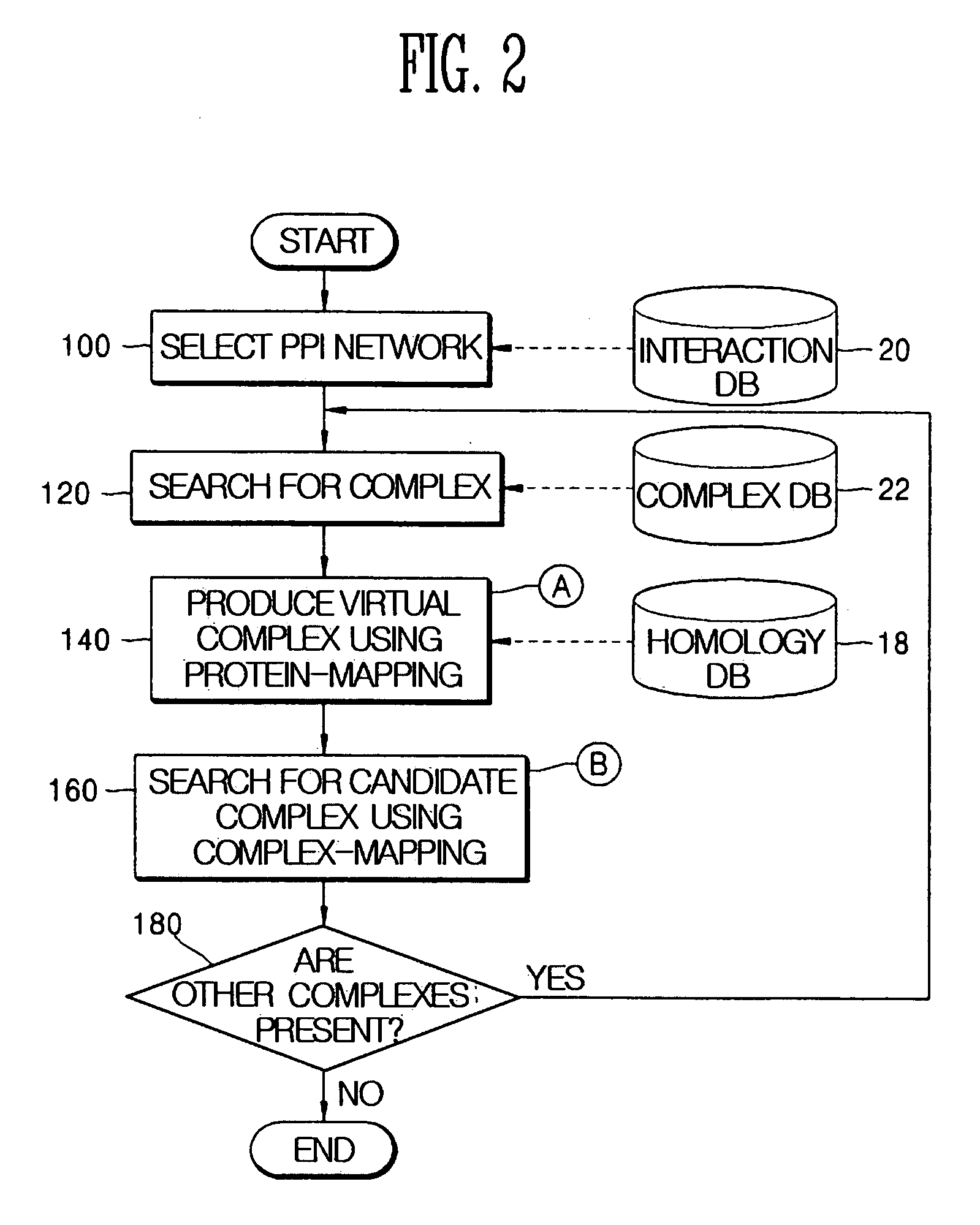
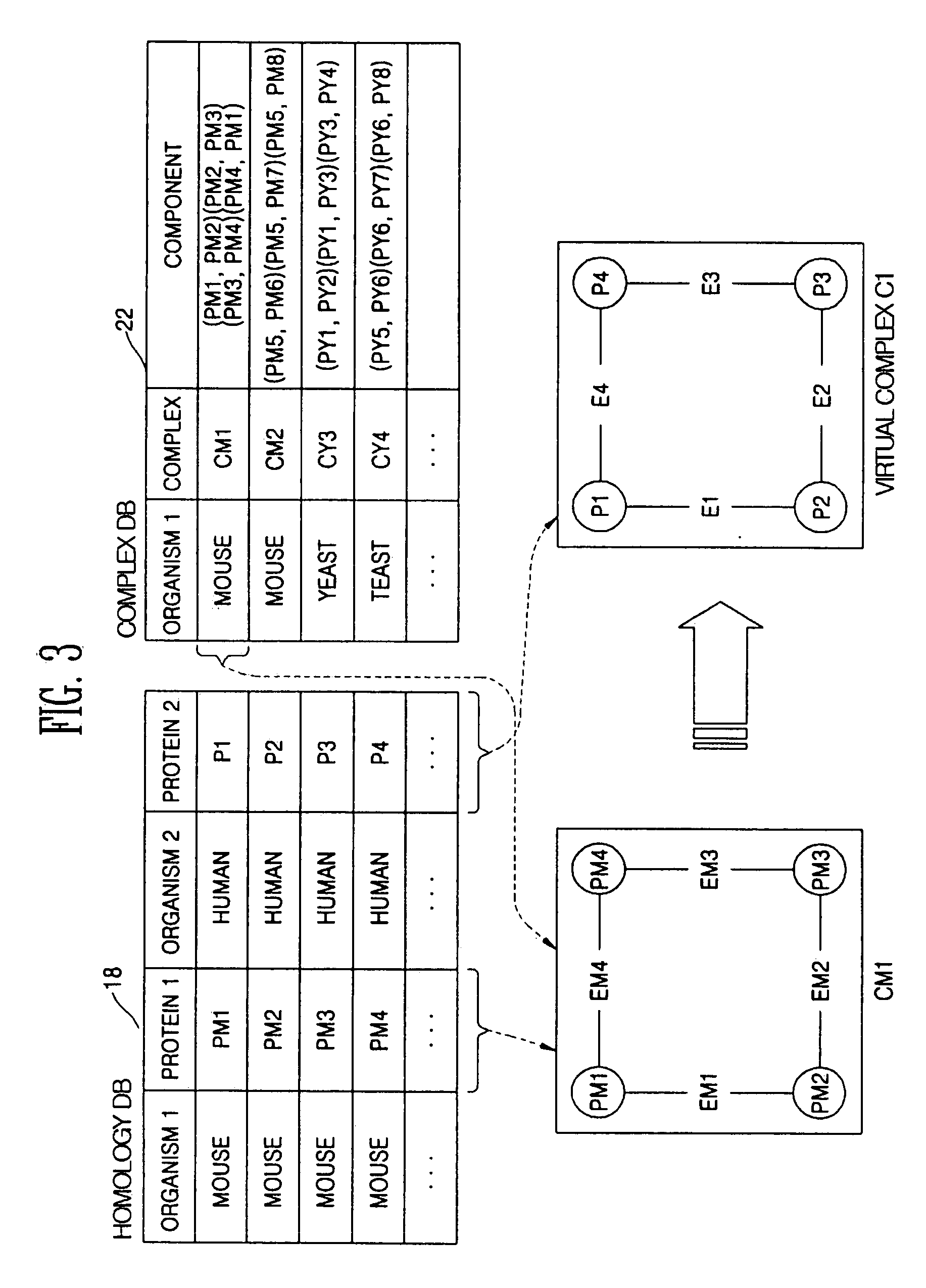
Method and apparatus for homology-based complex detection in a protein-protein interaction network
InactiveUS20060147999A1ProteomicsBiological testingProtein protein interaction networkProtein-protein complex
Provided are a method and a apparatus for detecting a protein complex using a similarity between different proteins in a protein-protein interaction network. The method includes: (a) producing a virtual complex of a specific organism by mapping proteins contained in a complex of a different organism into proteins of the specific organism using homology information between different proteins; and (b) searching for the produced virtual complex in the protein-protein interaction network of the specific organism.
Owner:ELECTRONICS & TELECOMM RES INST
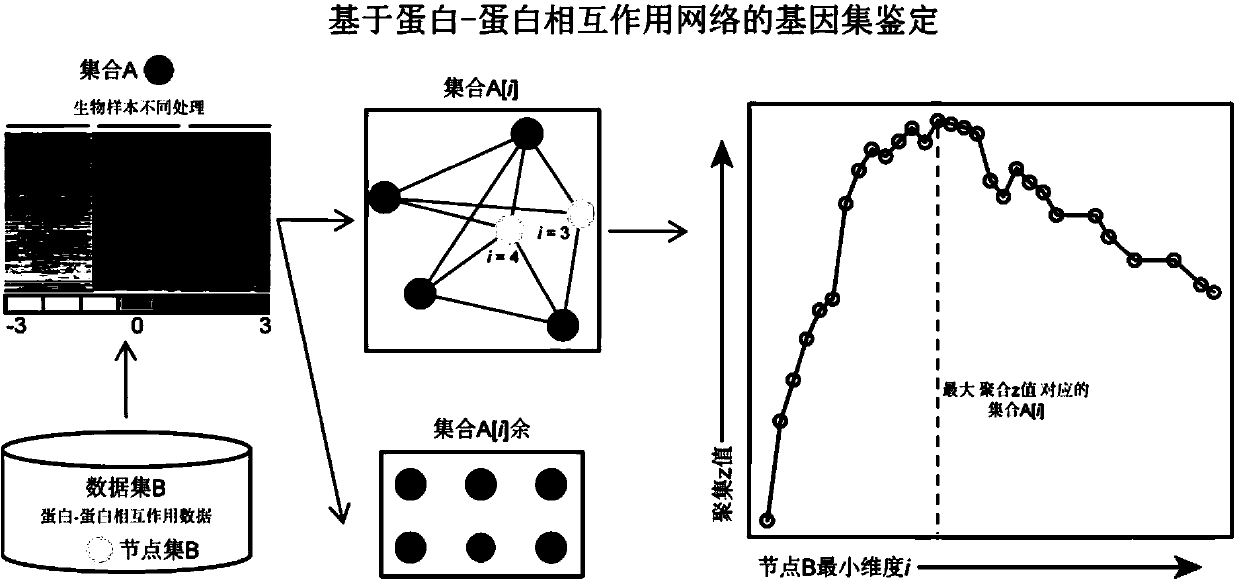
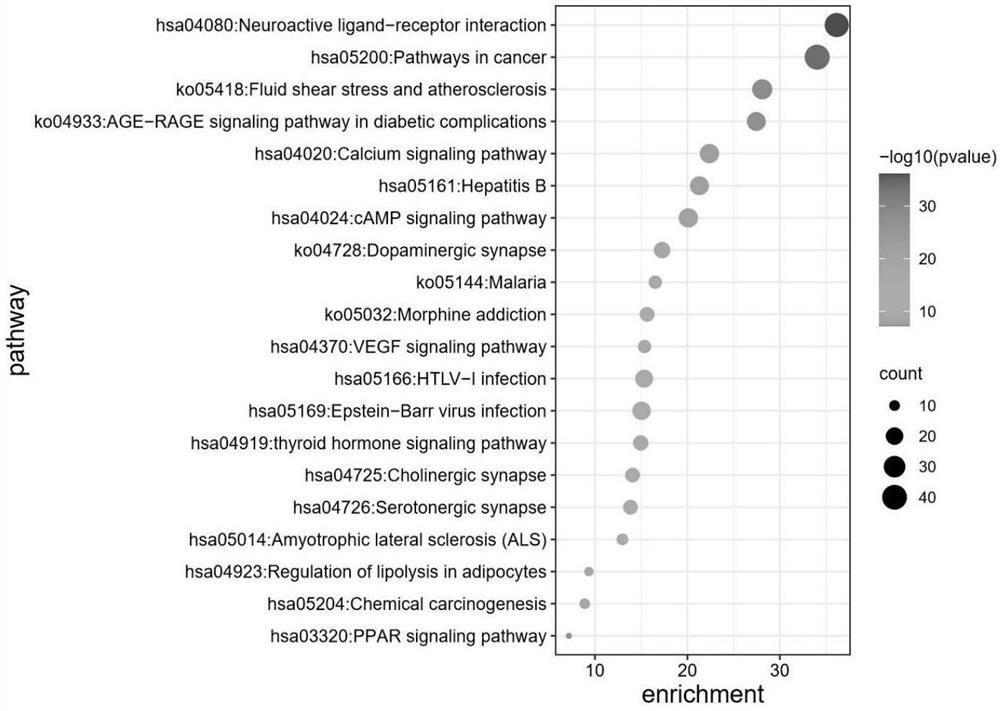
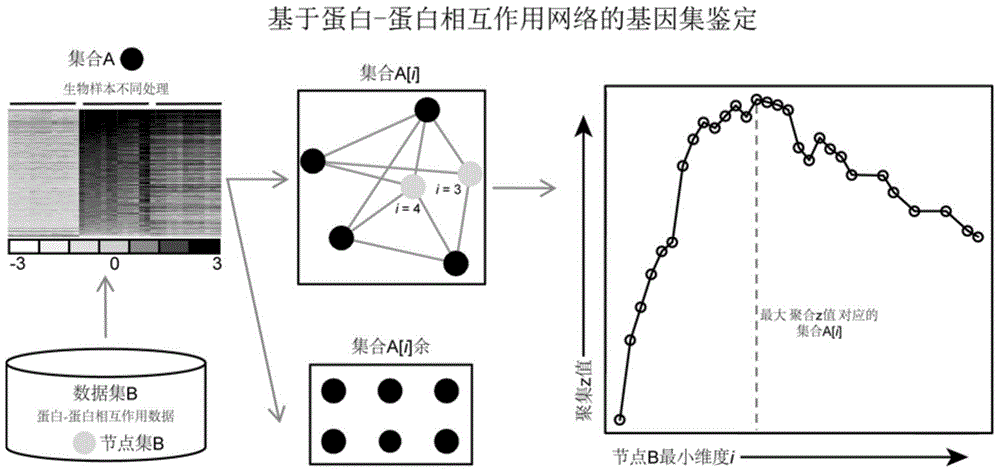
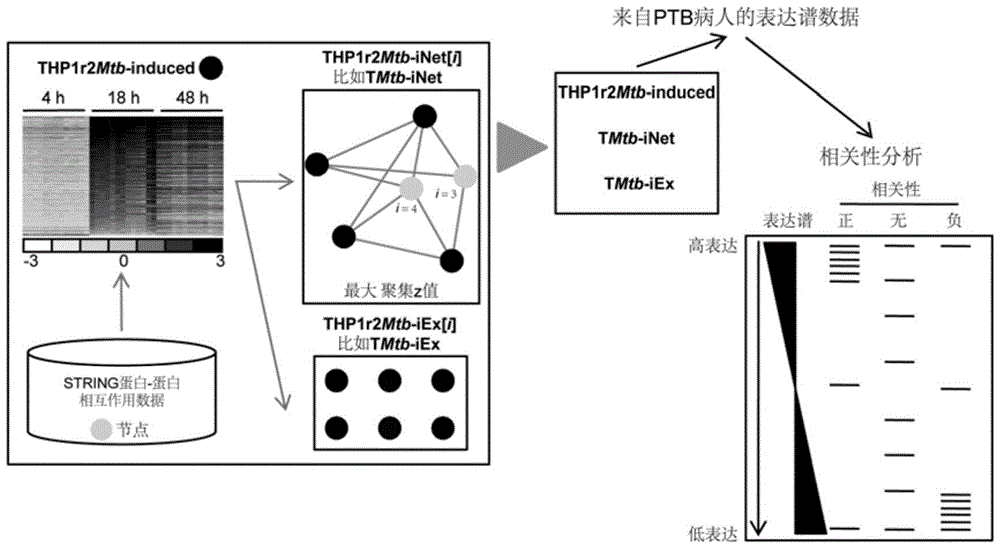
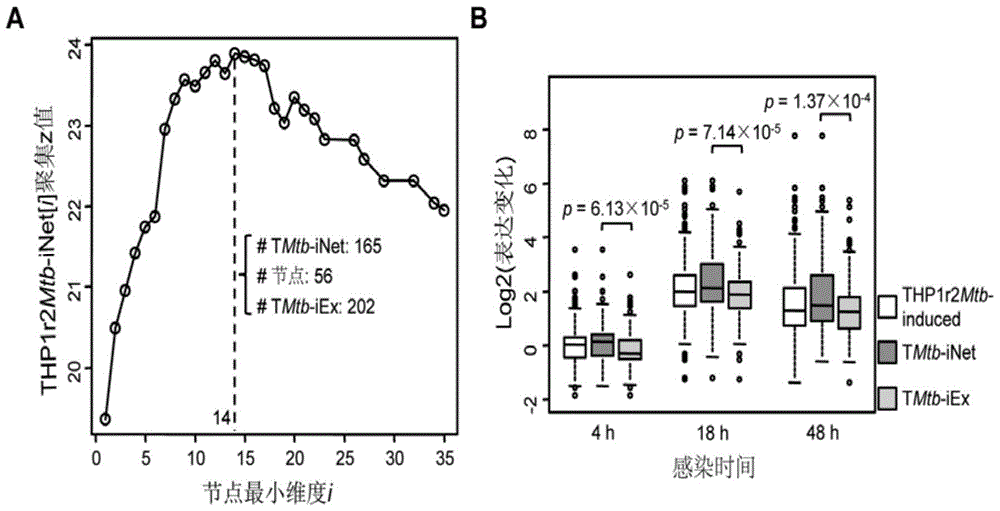
Protein-protein interaction network based gene set identification method
ActiveCN104182654ASignificant set modulationSpecial data processing applicationsProtein protein interaction networkProtein Interaction Networks
The invention relates to a protein-protein interaction network based gene set identification method, and belongs to the technical field of genes. The identification method comprises the following steps: finding out genes / proteins in direct interaction with a 'set A' from a 'set B', and naming as a 'node set B'; counting the number of each gene / protein in the 'node set B' in direct interaction with the 'set A', and naming as a dimensionality 'i'; calling out the interactive genes / proteins from the 'set A' through the 'node set B [i]' with different minimal dimensionalities 'i', and naming as a 'set A [i]'; counting the aggregate z-score of the 'set A [i]'; adopting the 'set A [i]' with the maximal aggregate z-score as the obtained gene set. The identification method can identify the gene sets more relevant to the biological processes, and is helpful for relevant researchers to carry out correlational research work.
Owner:SHANGHAI PUBLIC HEALTH CLINICAL CENT
A method for identifying key proteins in protein interaction networks
ActiveCN105279397BImprove recognition accuracySolve the costSpecial data processing applicationsProtein protein interaction networkTime-Consuming
The invention discloses a method for identifying key proteins in a protein interaction network. According to the protein interaction data, an undirected graph G is constructed, and the edge clustering coefficient of the graph is calculated. Compared with the existing technology, the invention considers protein Based on the topology characteristics of the interaction network, combined with gene expression profile data and gene function annotation information data, integrating three sets of data to predict key proteins can effectively reduce the impact of single data source data noise on prediction accuracy. The key protein characteristics reflected in the three types of data, the edge clustering coefficient of the interaction network, the Pearson correlation coefficient of the gene expression value, and the gene function similarity index, are combined to predict the key protein in the network, and the present invention can significantly improve the protein interaction network. The recognition accuracy of the key proteins in the medium, and a large number of key proteins can be predicted at one time, which solves the expensive cost and time-consuming problems of biological experiment methods.
Owner:EAST CHINA JIAOTONG UNIVERSITY
Starch-stop nerve-soothing decoction-autism core action gene target spot and screening method thereof
The invention discloses a start-stop nerve-soothing decoction-autism core action gene target spot and a screening method thereof.The screening method includes the steps that intersection target spots of active ingredients and diseases are obtained through database analysis of TCMSP, uniprot and the like, then a protein-protein interaction (PPI) network is constructed, and a result is visually analyzed through Cytoscape software to obtain important modules and core target spots in the PPI network; and carrying out gene GO and KEGG pathway enrichment analysis on the target spot by adopting Metascap, so as to obtain a visual graph of component-target spot-gene function-pathway-disease. The method solves the problems that the number of interactions between multiple active components and multi-target proteins is too large and screening is difficult, and has important application significance for guiding research and development of traditional Chinese medicine compounds.
Owner:AIR FORCE MEDICAL UNIV
Gene Set Identification Method Based on Protein-Protein Interaction Network
ActiveCN104182654BSignificant set modulationSpecial data processing applicationsProtein protein interaction networkProtein Interaction Networks
Owner:SHANGHAI PUBLIC HEALTH CLINICAL CENT
Predicting personalized cancer metastasis routes, biological mediators of metastasis and metastasis blocking therapies
InactiveCN109074425AReduce workloadLow costBiostatisticsMedical automated diagnosisProtein protein interaction networkCancer type
Predicting the metastasis of cancer in a patient from one tissue to another is disclosed. A computer-implemented method for predicting metastasis may comprise receiving an indication of at least one disrupted gene of the cancer; searching for data representing a gene-to-gene or protein-to-protein interaction network to determine the position of a received gene, wherein the data representing a gene-to-gene or protein-to-protein interaction network includes data that expresses the gene or the protein as a node of the network and expresses functional or physical interactions among the genes or proteins as an edge of the network; traversing the data representing a gene-to-gene or protein-to-protein interaction network specific for a type of the cancer type from a position of the received genein the network to a position of at least one gene involved in metastasis for a tissue type, organ or body part; determining at least one shortest path in the network between the received gene and theat least one gene involved in metastasis for the tissue type, organ or body part; generating a prediction of metastasis to the tissue type based on the at least one determined path; and generating anoutput display indicating a likelihood of spread of cancer to the tissue type, organ or body part.
Owner:IBM CORP
A Semantic Density-Based Method for Identification of Protein Network Complexes
InactiveCN104992078BReduce time complexityShort timeSpecial data processing applicationsNODALData set
The invention discloses a method for identifying protein network complexes based on semantic density, which is specifically implemented according to the following steps: for a weightless protein interaction network data set, search for the attributes of all proteins in the network data set in the gene ontology database GO; Based on the search results, a semantic similarity calculation method based on gene ontology is used to calculate the similarity between the connected proteins in the network dataset; according to the obtained similarity results, the given protein interaction network dataset is transformed into is a weighted, undirected network dataset, where nodes represent proteins, edges represent interactions between proteins, and the similarity between proteins is the weight of the edges; protein complexes can be identified from the protein interaction network, and The recognition accuracy is higher and the time complexity is lower.
Owner:XIAN UNIV OF TECH
A method and system for determining cancer network markers based on a probability model
InactiveCN109101783BGet accurate and effectiveAccurate diagnosisSystems biologyInstrumentsDiseaseProtein protein interaction network
The invention discloses a method and a system for determining cancer network markers based on a probability model. The method comprises the following steps: utilizing a probability density function, converting gene expression data matrices of all normal samples and disease samples obtained into likelihood matrices, and constructing a normal sample distribution function according to the likelihoodmatrices of all normal samples; each element in the likelihood matrix of each disease sample is then brought into the normal sample distribution function, the set of significantly different genes foreach disease sample is determined, and the set of significantly different genes for each disease sample is mapped to proteins. Protein-protein interaction networks, and network markers for each disease sample are identified. The method or system provided by the invention can accurately and effectively acquire cancer network markers, and utilize the cancer network markers to carry out subtype classification of diseases so as to realize precise diagnosis and treatment of diseases.
Owner:WENZHOU UNIVERSITY
System and method for determining an association of at least one biological feature with a medical condition
InactiveUS10192642B2Medical simulationBiostatisticsProtein protein interaction networkProtein Interaction Networks
A system and a method for determining an association of at least one biological feature with a medical condition, in particularly, but not exclusively, a system and a method of determining an association of at least one biological feature in form of a gene expression with cancer or a subtype of cancer that can include the generation of a simplified protein-protein interaction network based on processed biological data. The system and respective method is especially suitable for analysis of high dimensional and low sample size biological datasets such as in cancer research.
Owner:MACAU UNIV OF SCI & TECH
Identification of protein complexes using Drosophila-optimized methods
InactiveCN105868582BImprove accuracyRealistically simulate dynamicsProteomicsGenomicsUndirected graphProtein protein interaction network
Owner:SHAANXI NORMAL UNIV
Analytical method and application of fragmentable chemical cross-linking agent based on glycosidic bond mass spectrometry
ActiveCN109900814BLow trait requirementsFast analysisComponent separationProtein containing complexProtein protein interaction network
The present invention relates to an analysis method based on glycosidic bond mass spectrometry fragmentable chemical cross-linking agent for analyzing protein-protein interaction by chemical cross-linking mass spectrometry. The chemical cross-linking reaction between the cross-linking agent and the protein complex is carried out, and the selective difference in the fragmentation energy of the glycosidic bond and the peptide bond is used to realize the highly selective and controllable fragmentation of the cross-linking reagent and the peptide bond of the cross-linked peptide, thereby Reduce the complexity of the spectrum of cross-linked peptides and significantly reduce the scale of data retrieval, realize the large-scale analysis of protein complexes based on chemical cross-linking strategies, and provide an important technology for studying the spatial structure of proteins and protein-protein interaction networks support.
Owner:DALIAN INST OF CHEM PHYSICS CHINESE ACAD OF SCI
A method for identifying key proteins using an artificial bee colony optimization algorithm using a foraging mechanism
InactiveCN106874708BFeaturesSolve the shortcomings of not being able to consider the overall nature of the networkArtificial lifeProteomicsProtein protein interaction networkPerformance index
The invention discloses a method for identifying key protein using an artificial bee colony optimization algorithm of a foraging mechanism. The method comprises: converting a protein-protein interaction network to an undirected graph, obtaining ribonucleic acid genetic expression values corresponding to protein, preprocessing edges and nodes of the protein-protein interaction network, establishing a dynamic protein-protein interaction network, selecting known key protein as a honey source, honey bees searching neighbourhood of the honey source, following bees searching neighbourhood of the honey bees, updating the honey sources, investigating bees searching new honey sources in a global manner, updating the honey sources, and generating the key protein. The method can accurately identify the key protein. Results of simulation experiments show that sensitiveness, specificity, positive predictive values, negative predictive values and other performance indexes are relatively excellent. Compared with other key protein identification method, the identification process of key protein realized by combining optimizing characteristics of artificial bee colony with characteristics of the protein-protein interaction network improves identification accuracy rate of the key protein.
Owner:SHAANXI NORMAL UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com