A method of identifying protein compounds by using a fruit fly optimization method
A protein complex and Drosophila optimization technology, applied in the field of bioinformatics, can solve the problem of low recognition accuracy of protein complexes, the inability to consider the internal structure of protein complexes with global characteristics and local characteristics, and failure to consider protein interactions Network characteristics and other issues to achieve high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
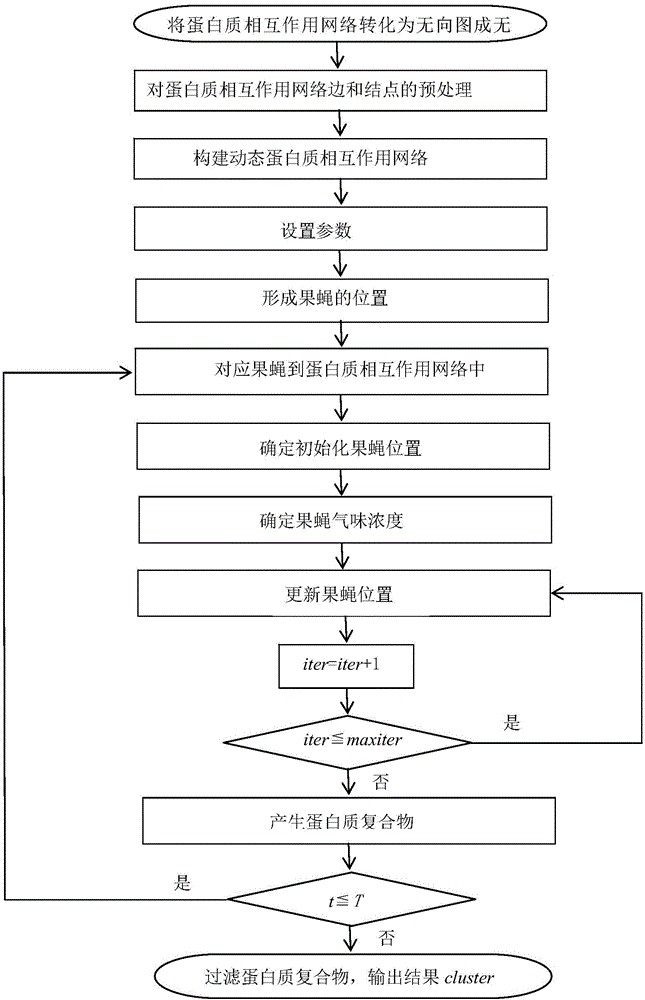
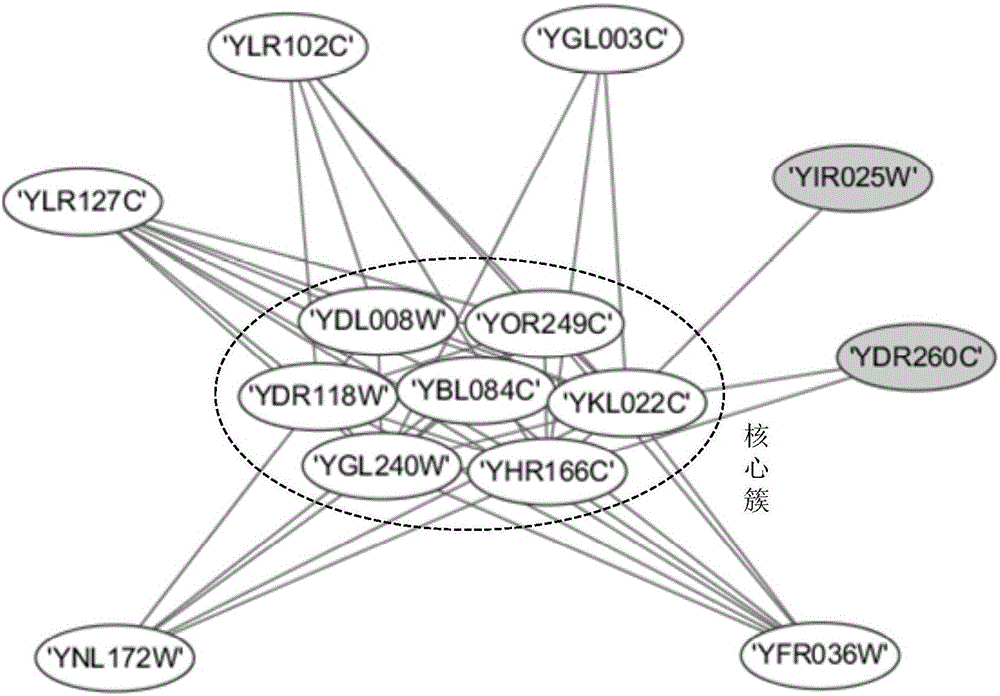
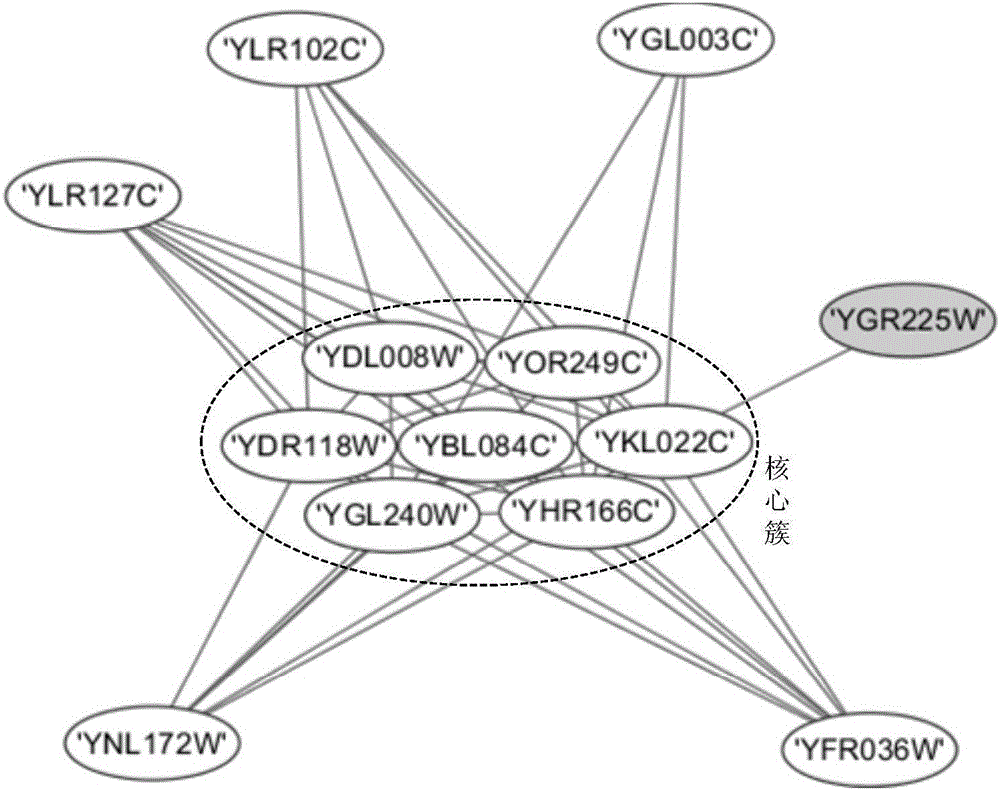
[0064] Taking 12 dynamic protein networks as an example, the steps to identify protein complexes using the Drosophila optimization method are as follows:
[0065] In this embodiment, the yeast data set (DIP 20140427 version) collected from the DIP database is used as the simulation data set. The DIP data contains 4995 proteins and 21554 interaction relationships. The gene expression dataset is collected from the yeast metabolic expression dataset GSE3431 in the GEO database, which includes 6777 genes, gene values at 36 time points in 3 cycles, covering 95% of the proteins in DIP. Using gene expression values to create 12 dynamic protein interaction networks. The experimental platform is Windows 7 operating system, Intel Core 2 Duo 3.1GHz processor, 4GB physical memory, and realizes the FOCA method of the present invention with Matlab R2010b software.
[0066] 1. Transform the protein interaction network into an undirected graph
[0067] Transform the protein interaction ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com