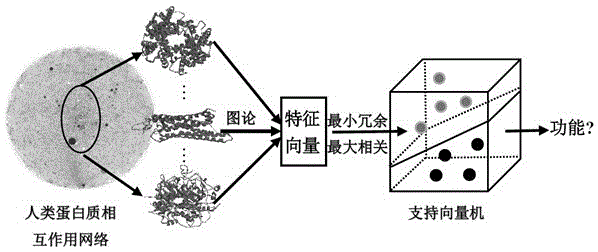
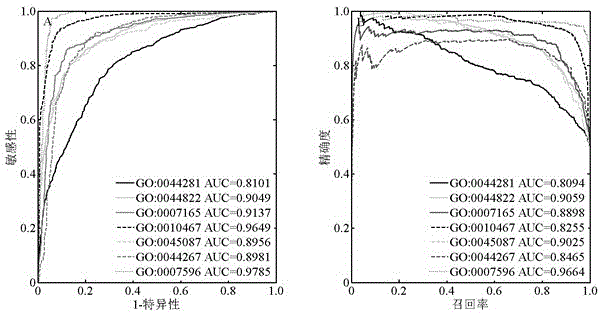
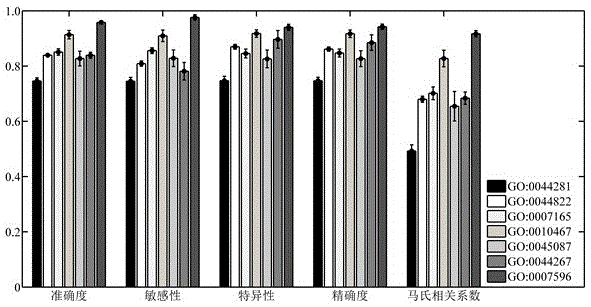
Method for identifying protein functions based on protein-protein interaction network and network topological structure features
A technology of network topology and protein function, applied in the field of proteomics, can solve the problems of changing the biological function of protein, not taking into account the different biological functions of homologous proteins, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] 1. Collect data sets and construct protein interaction networks weighted by nodes and edges
[0059] Human protein-protein interaction data were collected from the HIPPIE database, removing self-interactions, repeated interactions, and interactions with an interaction score of 0. According to the protein acquisition number, the protein primary structure data was obtained from the UniprotKB / Swiss-Prot database, and the amino acid composition, dipeptide composition, autocorrelation descriptors and protein primary structure descriptors such as composition, transformation and distribution were calculated. A protein-protein interaction network weighted by nodes and edges is constructed. Nodes represent proteins, edges represent interactions, node weights are the primary structural features of proteins, and edge weights are interaction trust scores.
[0060] 2. Collect protein function annotation data and build a data set
[0061] Proteins with molecular function and biologi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com