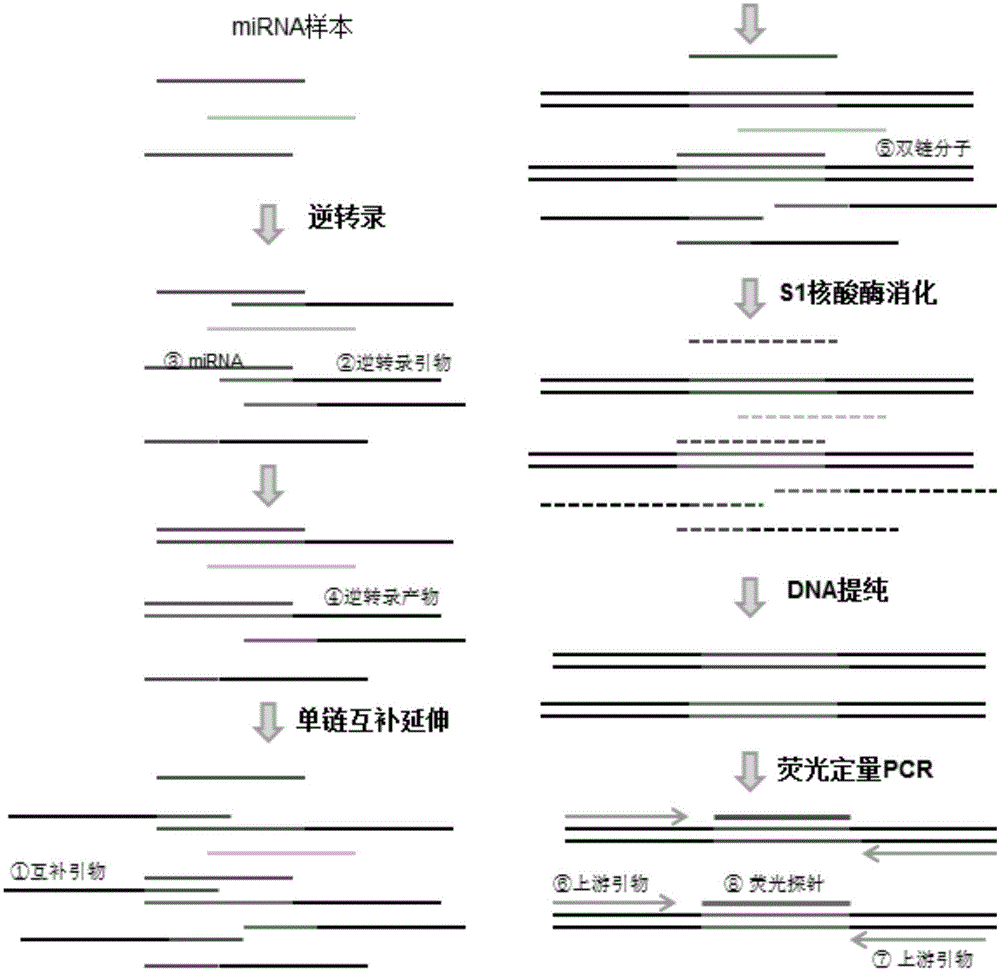
Method for conducting multiplex quantitative detection on miRNA
A quantitative detection and multiplexing technology, applied in the field of fluorescence quantitative PCR, which can solve the problems of low sensitivity and low repeatability of detection results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
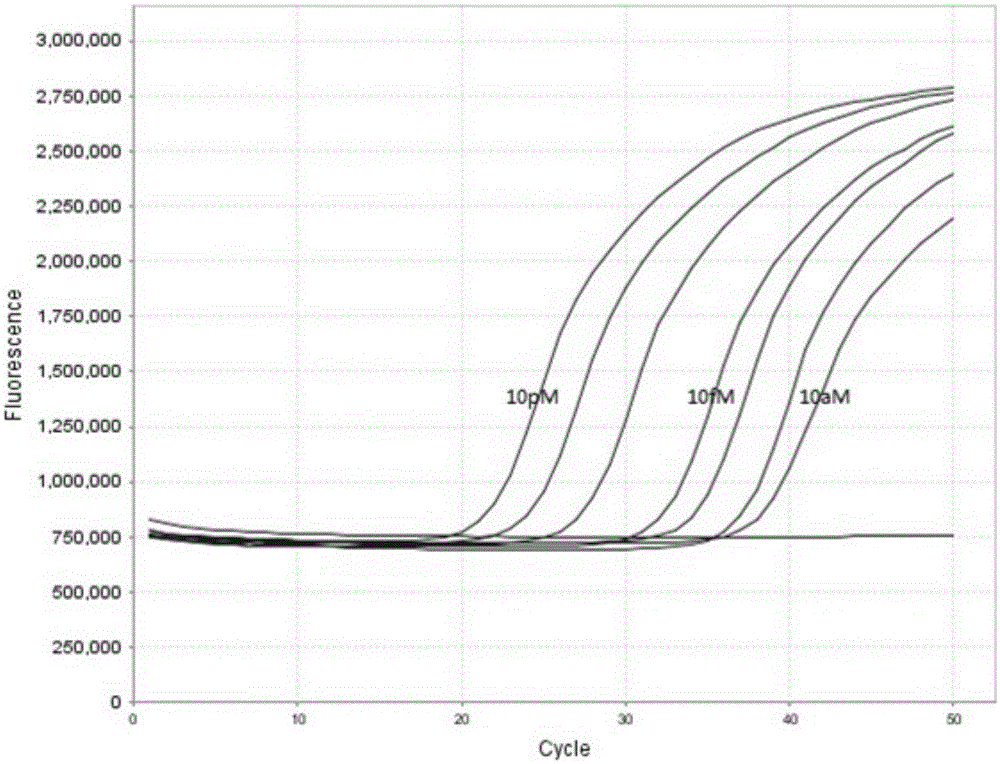
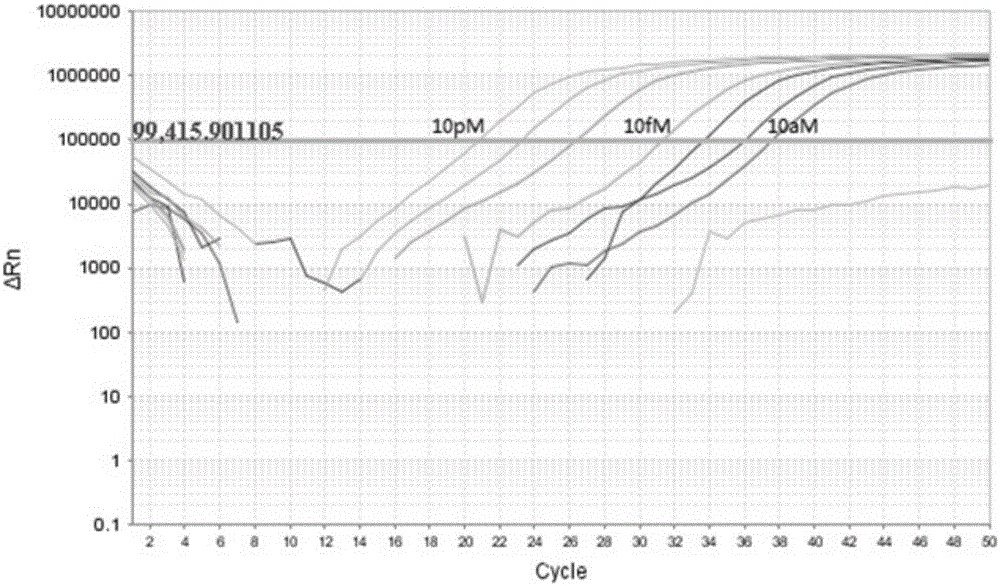
[0052] Example 1, Quantitative detection of two miRNA examples
[0053] 1. Tested miRNA
[0054] In this example, miR-16 and miR-39 were quantitatively detected simultaneously by the method of the invention described above. miR-16 is a miRNA that is continuously expressed in the human body, and its expression level has a certain relationship with some diseases. miR-39 is an miRNA that is expressed in nematodes but not in humans, so it is often used as a quality control and added to samples. Simultaneous detection of miR-16 and miR-39 helps to standardize the detection of miR-16.
[0055] 2. Sequence design
[0056] (1) The sequences of miR-16 and miR-39 were obtained according to the database as follows:
[0057] miR-16: 5'-UAGCAGCACGUAAAUAUUGGCG-3' (SEQ ID NO: 1);
[0058] miR-39: 5'-UCACCGGGUGUAAAUCAGCUUG-3' (SEQ ID NO: 2)
[0059] (2) Reverse transcription primers, complementary primers and hydrolysis probes for miR-16 and miR-39 were designed as follows:
[0060] Re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com