Reference genome and de novo assembly combination based next-generation sequencing data assembly method
A reference genome and next-generation sequencing technology, applied in electrical digital data processing, special data processing applications, instruments, etc., can solve the problems of assembly integrity discount, large impact, low quality, etc., to reduce complexity and improve continuity. , the effect of improving continuity and accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
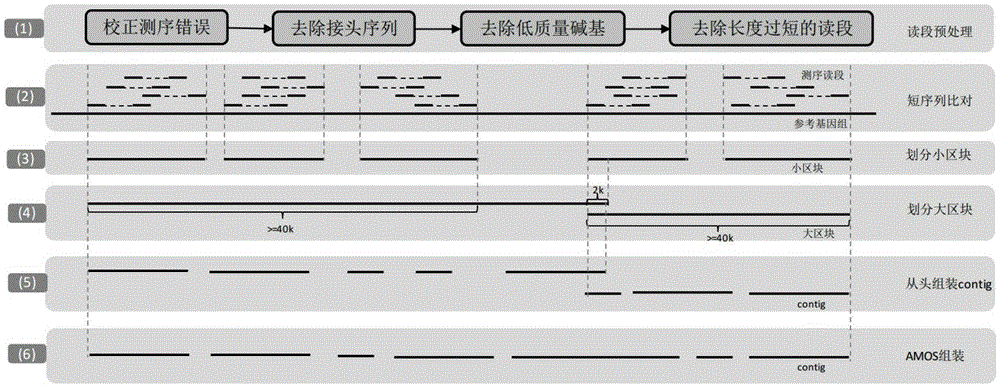
[0048] In this study, the sequencing data used were the whole-genome shotgun sequencing data of leaves of MH63 and ZS97 (indica rice varieties Minghui 63 and Zhenshan 97) provided by the rice research team of Huazhong Agricultural University. The sampling period was the three-leaf stage of rice, the sequencing platform IlluminaHiseq2000, PE100 sequencing, a total of 300bp, 5k, 10k libraries of three different insert-size (Table 1). In addition, the Nipponbare Genome Reference Genome IRGSP-1.0 (http: / / rapdb.dna.affrc.go.jp / ) was also used
[0049] Table 1 Sequencing data statistics
[0050]
[0051] a According to the read alignment to Nipponbare genome statistics.
[0052] b Based on Nipponbare genome size estimates. .
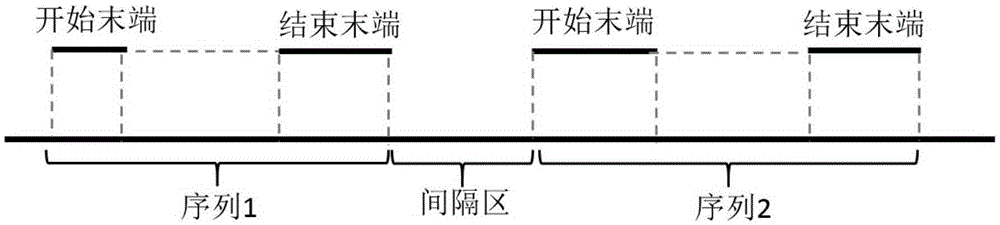
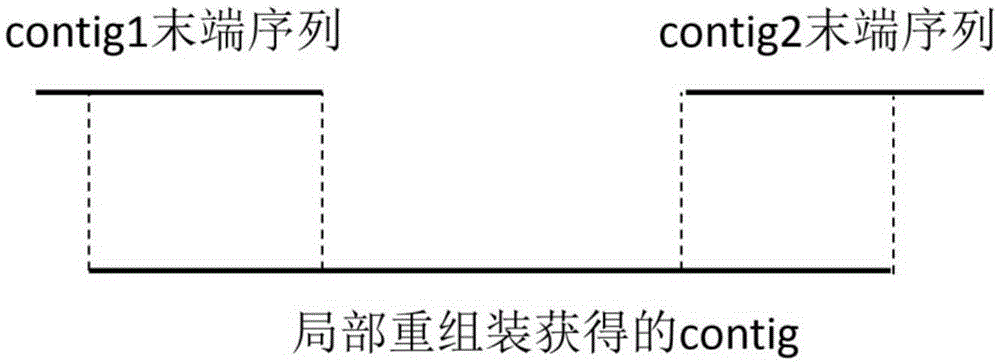
[0053] We adopted a strategy of assembling based on the Nipponbare reference genome. We divided multiple regions based on the Nipponbare sequence, and performed partial de novo assembly in each region. The Nipponbare sequence, whole genome de novo as...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com