Specific biomarker set for non-invasive diagnosis of liver cancer
A biomarker and specific technology, applied in disease diagnosis, biological testing, drug combination, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1a
[0064] Protein extraction from patient biopsies
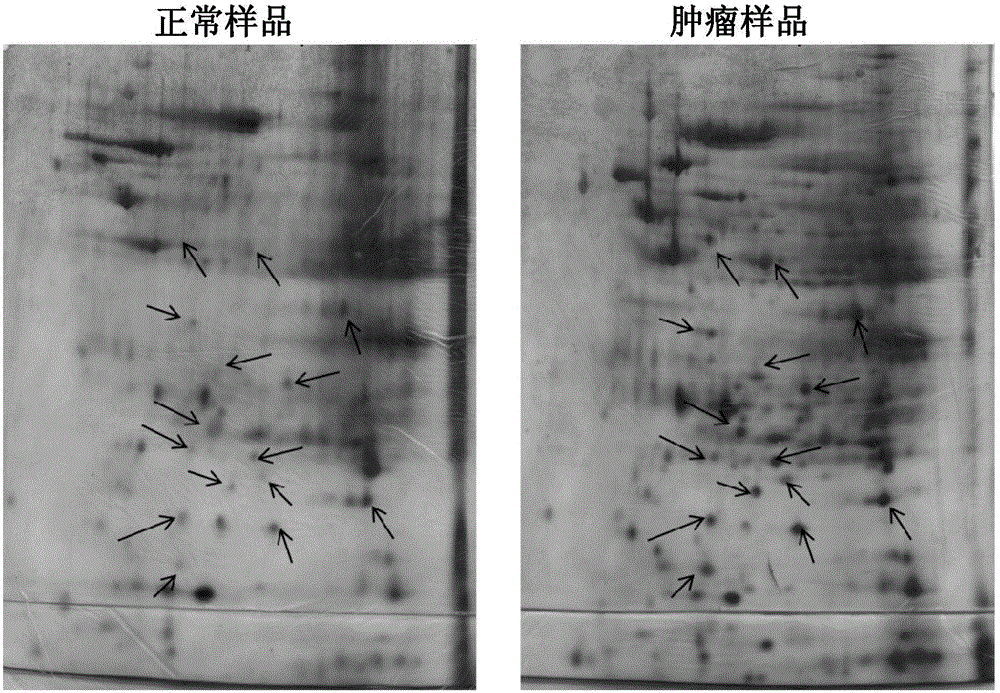
[0065] Paired biopsies (tumor biopsies versus adjacent normal tissue) of 500 mg patients were collected and washed with PBS. Freeze the tissue by submersion in liquid nitrogen and immediately homogenize the tissue with a pestle and mortar. To the homogenized sample, a lysis solution (8M urea, 4% CHAPS, 2% IPG buffer, 0.2 mg / ml PMSF) was added, followed by vortexing for at least 5 min until the tissue was completely dispersed. The lysate was then clarified by centrifugation at 14,000 rpm for 10 minutes at 4°C. The supernatant was further washed by 2DCleanUpkit (Amersham) to remove salts and impurities. Resuspend the pellet with minimal volume of rehydration solution (without addition of DTT & IPG buffer). Protein concentrations were then measured by the Bio-Rad protein assay and aliquots of 200 g / tube were stored at -70°C.
Embodiment 1b
[0067] Protein Analysis by 2D Electrophoresis
[0068] To 1 ml rehydration stock solution was added 2.8 mg DTT, 5 μl pharmalyte or IPG buffer and 2 μl bromophenol blue. 50-100 μg protein sample was added to a 13 cm Immobiline Dry Strip (IPG strip) containing 250 μl rehydration solution. After removing the protective cover, position the IPG strips in the strip rack, gel side down, and cover with CoverFluid to prevent dehydration during electrophoresis. The strips were then placed on an Ettan IPGphor (Amersham) for isoelectric focusing (first dimension electrophoresis).
[0069] After the first dimension electrophoresis, the IPG strips were equilibrated with equilibration solution (6M urea 2% SDS, 50mM TrisHCl pH6.8, 30% glycerol, 0.002% bromophenol blue, 100mgDTT per 10ml buffer and 250mgIAA per 10ml buffer), and then used Wash 4–5 times with 1xSDS electrophoresis buffer. The IPG strip was placed on top of the second dimension gel and covered with sealing solution (0.5% low ...
Embodiment 2a
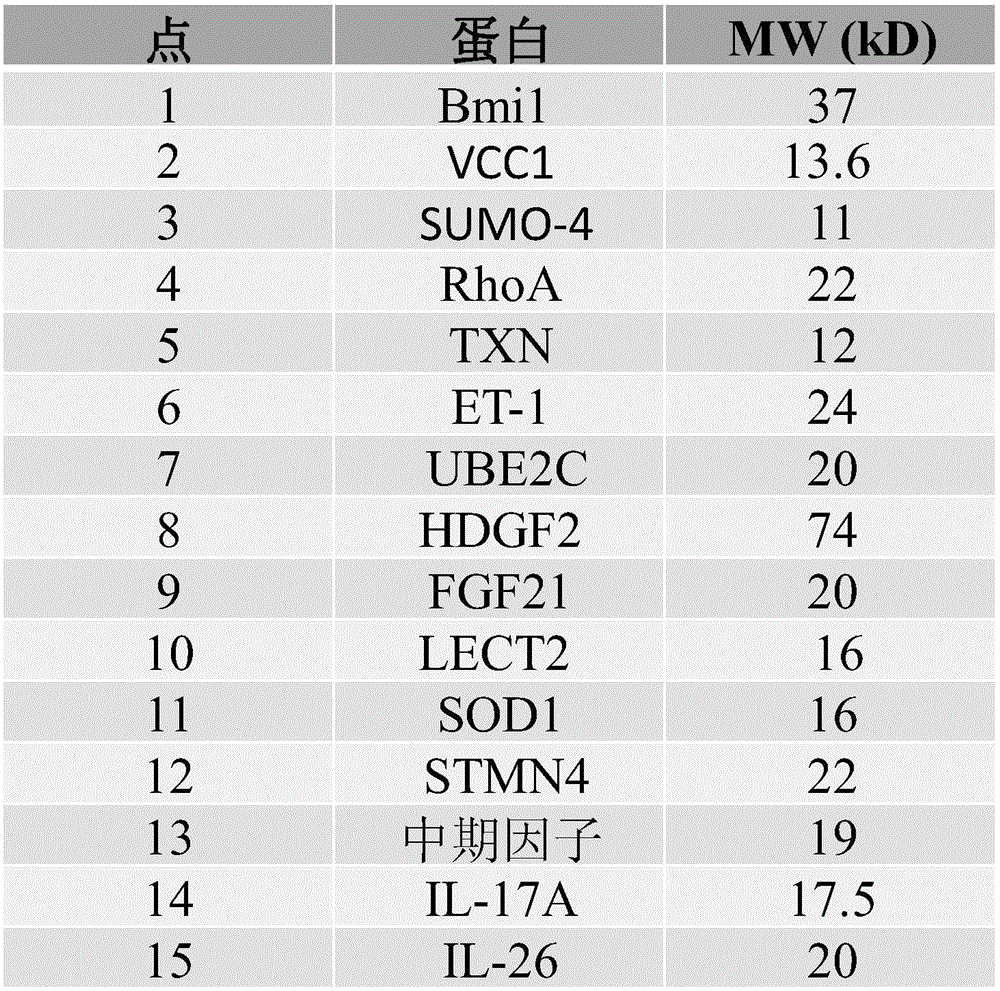
[0071] Example 2a (SEQ ID NO.1)
[0072] Amino acid sequence of Bmi1
[0073] MHRTTRIKITELNPHLMCVLCGGYFIDATTIIECLHSFCKTCIVRYLETSKYCPICDVQVHKTRPLLNIRSDKTLQDIVYKLVPGLFKNEMKRRRDFYAAHPSADAANGSNEDRGEVADEDKRIITDDEIISLSIEFFDQNRLDRKVNKDKEKSKEEVNDKRYLRCPAAMTVMHLRKFLRSKMDIPNTFQIDVMYEEEPLKDYYTLMDIAYIYTWRRNGPLPLKYRVRPTCKRMKISHQRDGLTNAGELESDSGSDKANSPAGGIPSTSSCLPSPSTPVQSPHPQFPHISSTMNGTSNSPSGNHQSSFANRPRKSSVNGSSATSSG
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com