Gene order fragment fast positioning method based on bitmap
A gene sequence and positioning method technology, applied in genomics, special data processing applications, instruments, etc., can solve problems such as computing burdens, achieve fast positioning, accurate positioning, and accelerate the organization of data query processes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
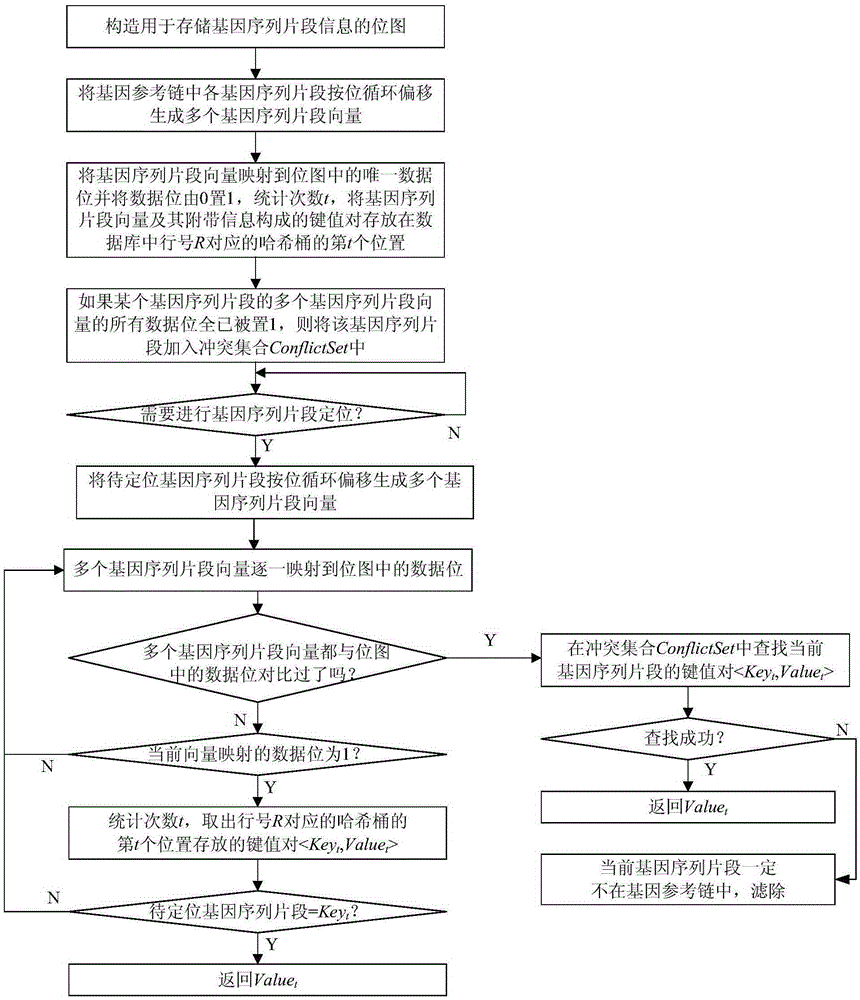
[0037] Such as figure 1 As shown, the steps of the method for rapidly locating gene sequence fragments based on bitmaps in this embodiment include:
[0038] 1) Construct a bitmap for storing gene sequence fragment information, and each data bit of the bitmap is initialized to 0;
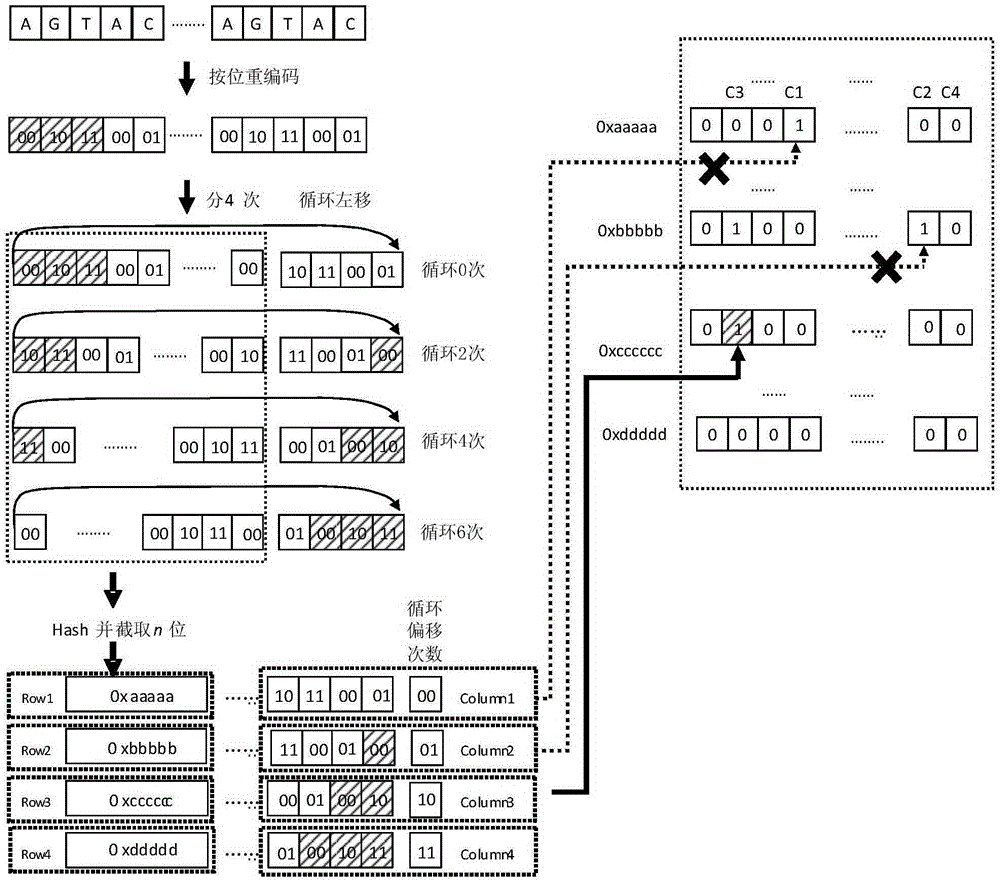
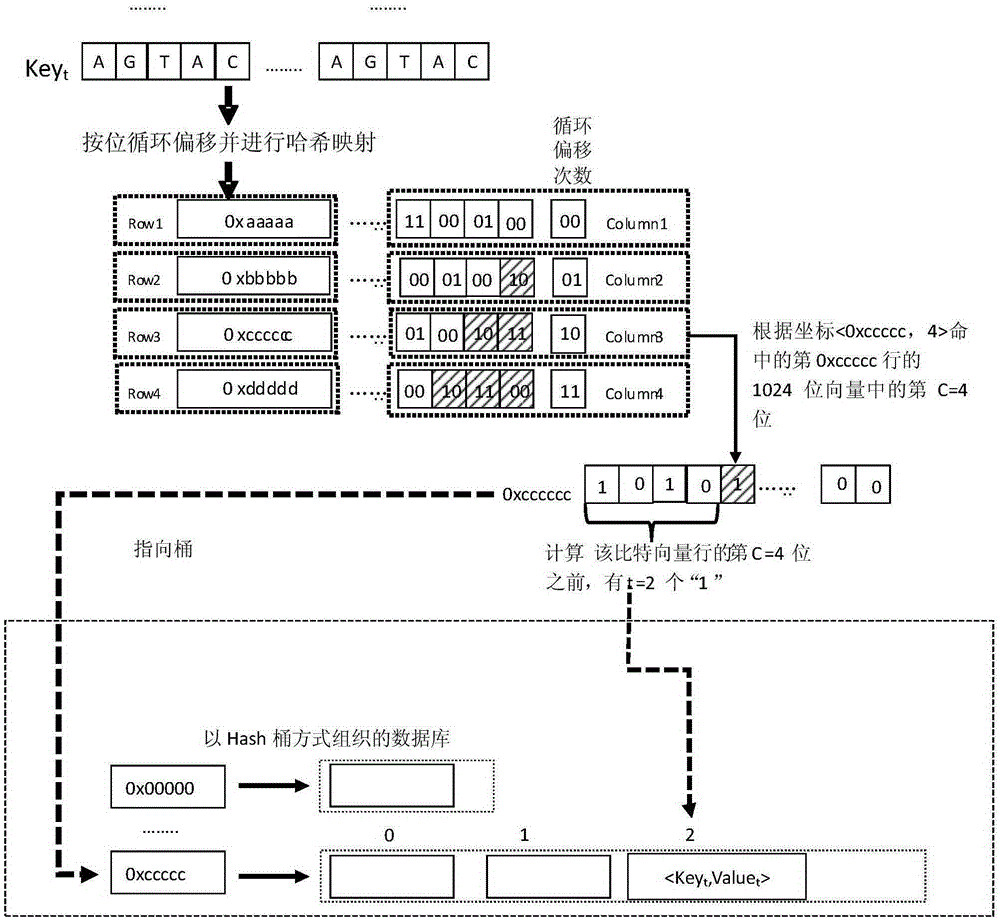
[0039] 2) The gene sequence fragments in the gene reference chain are cyclically shifted to generate a plurality of gene sequence fragment vectors, and the gene sequence fragment vectors are mapped to the unique data bits in the bitmap using a hash function and the data bits are set from 0 to 1, Count the number of occurrences t of all data bits in the row R where the data bit is located from the 0th column to the column where the data bit is located, and the key-value pair t ,Value t >Store in the tth position of the hash bucket corresponding to the row number R of the data bit in the database; if all the data bits of the multiple gene sequence segment vectors of a certain gene sequence segment hav...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com