Method and kit for detecting ribonuclease
A ribonuclease and ribonucleotide technology, which is applied in the field of a method for detecting ribonuclease and a kit thereof, can solve problems such as the inability to meet the needs of routine quality control in laboratories, and achieves low cost, high sensitivity, The effect of small reaction volume
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Embodiment 1, be used to detect the RNA probe synthesis of ribonuclease
[0069] 1. Design principles of RNA probes
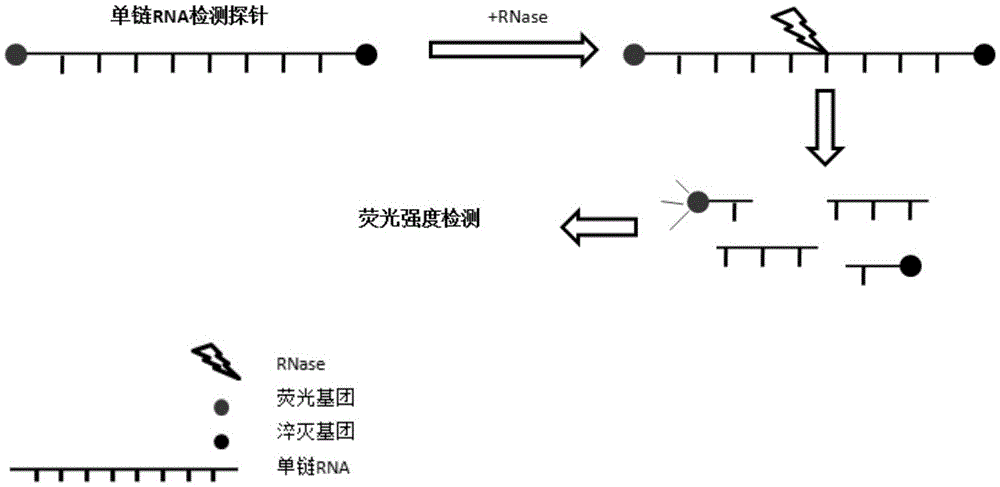
[0070] 1) The RNA probe consists of a single-stranded RNA, an energy donor group and an energy acceptor group;
[0071] Single-stranded RNA consists of 2-30 ribonucleotides, preferably 4-10 ribonucleotides; single-stranded RNA specifically consists of 4, 5, 6, 9 or 10 nucleotides.
[0072] The single-stranded RNA comprises at least one ribonuclease cleavage site, at least 1 G and at least 1 C, preferably the single-stranded RNA comprises at least one ribonuclease cleavage site and at least 1 GC sequence. Preferably, the nucleotide next to the G of the GC sequence or the CG sequence is A or U;
[0073] The energy donor group and the energy acceptor group are connected to the nucleotides at the two ends of the single-stranded RNA, the energy donor group is a fluorescent emitting group, and the energy acceptor group is a fluorescent quencher corresponding...
Embodiment 2
[0082] Embodiment 2, detect RNaseA
[0083] 1. Reagents for detection
[0084] 1. Preparation of RNA probes and reagents
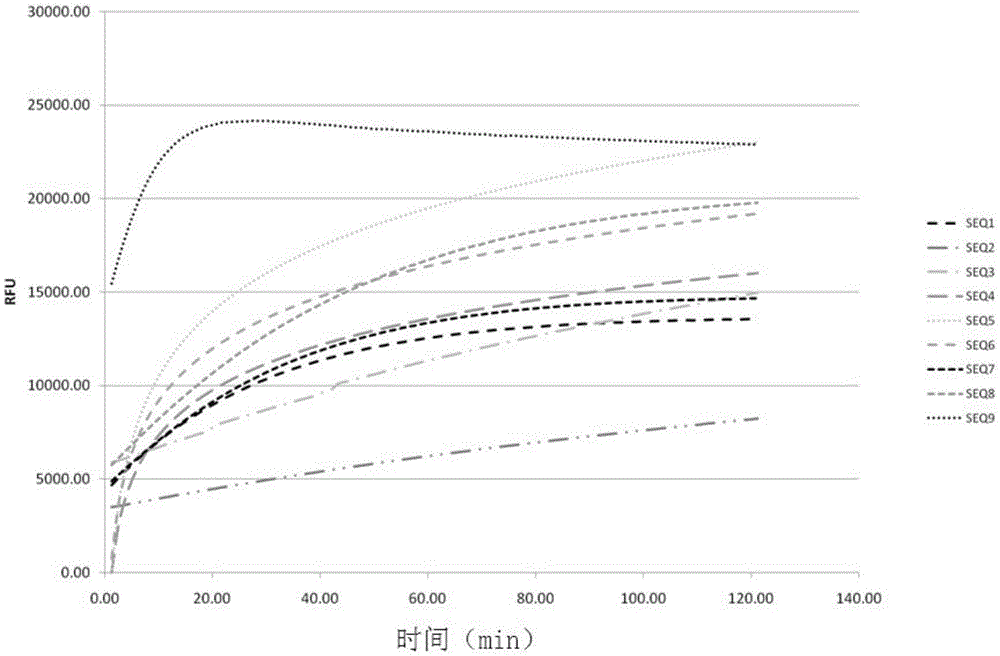
[0085] The RNA probes shown in Table 1 were synthesized.
[0086] 2. RNase probe preparation
[0087] RNase probe stock solution preparation (100pmol / ul): RNA probe dry powder added RNasefreeddH 2 O, make the concentration of the RNA probe 100 pmol / ul, obtain the stock solution, and store it in the dark at -20°C.
[0088] RNase probe working solution preparation (10pmol / ul): take RNase-free 1.5ml tube, add 90ulddH 2 O and 10ul RNase probe stock solution, mix well, and store at -20°C in the dark.
[0089] 3. Preparation of 10× reaction buffer
[0090] Table 2 for 10x reaction buffer
[0091] components
scope
specific value
NaCl
0-5M, and not 0
50mM
KCl
0-5M, and not 0
20mM
20mM-1M
20mM
MgCl 2
10-100mM
10mM
pH
6.5-9.0
8.0
[0092]Prepare a 10× r...
Embodiment 3
[0107] Embodiment 3, the sensitivity research of RNA probe detection RNaseA
[0108] 1. Preparation of reaction system
[0109] Configure according to Table 2 and Table 3 of Example 2, wherein the RNA probe is the SEQ9 probe to obtain the reaction system.
[0110] 2. Detection
[0111] The samples to be tested are RNaseA diluted with water according to different dilution factors.
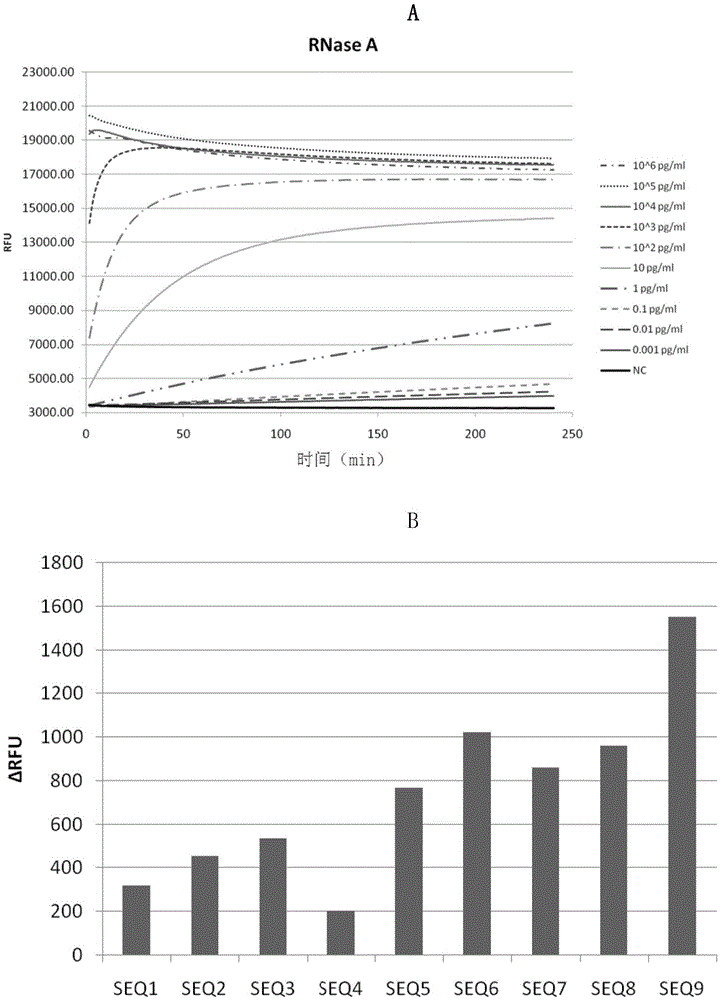
[0112] Add the above-mentioned RNaseA diluted with water at different dilution times to the reaction system of different tubes, so that the concentration of RNaseA in the reaction system is 10 6 pg / ml, 10 5 pg / ml, 10 4 pg / ml, 10 3 pg / ml, 10 2 pg / ml, 10pg / ml, 1pg / ml, 0.1pg / ml, 0.01pg / ml, 0.001pg / ml.
[0113] Detection method is the same as that of Example 2.
[0114] The result is as image 3 A, shows that adopting the detection method of the present invention, by using SEQ9 to detect RNaseA of different concentrations, the lower limit of detection of RNaseA is 0.1 pg / ml.
[0115] Will im...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com