A method for discovering specific functional antibodies
A specific, antibody technology, applied in chemical instruments and methods, botanical equipment and methods, biochemical equipment and methods, etc., can solve the problems of difficult bias, difficult samples, inaccurate antibody gene spectrum information, etc. Low, high throughput, simplified build process effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
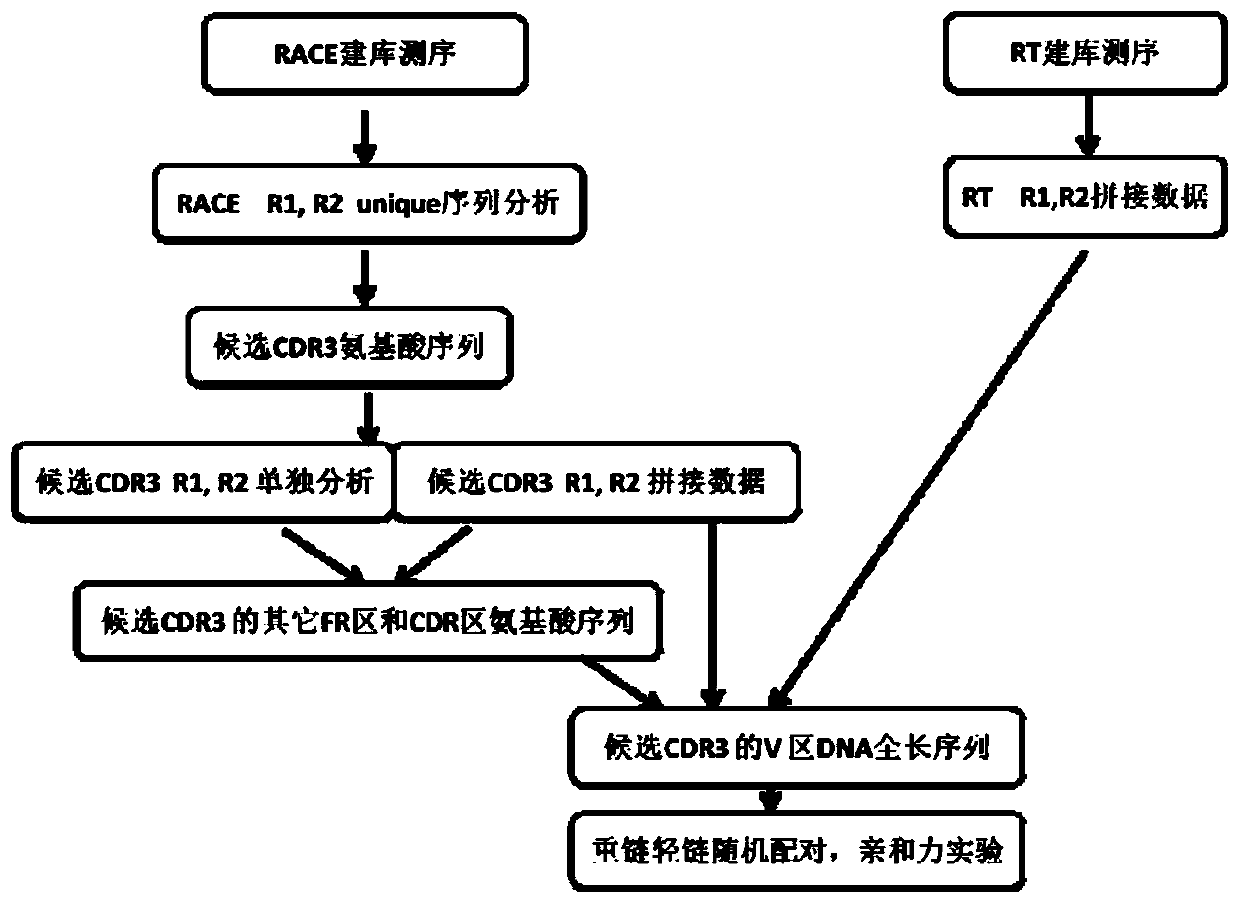
[0084] Example 1 An improved high-throughput library construction method suitable for micro-sample
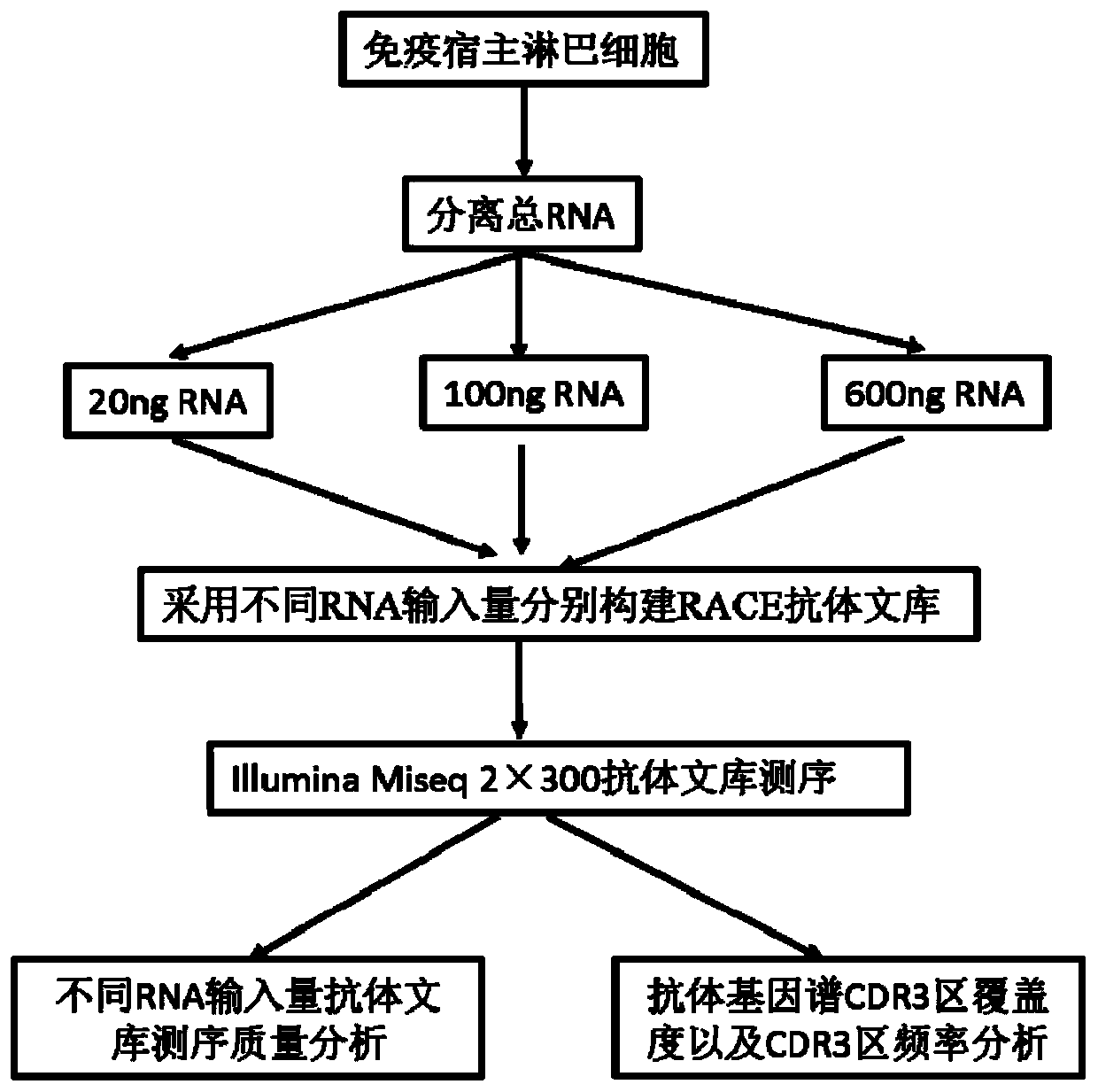
[0085] The specific steps of library construction are as follows: figure 2 As shown, sample preparation and first-strand cDNA synthesis: Peripheral blood mononuclear cells (PBMC) isolated from human or animal peripheral blood by density gradient centrifugation. Total RNA was isolated from PBMC using Trizol. Take 20ng, 100ng and 600ng of RNA as the template and oligo(dT) as the primer for the synthesis of the first-strand cDNA. For specific operations, refer to the kit RACE 5' / 3'Kit instructions.
[0086] The optimized two-step PCR method constructs the antibody library, the first round of PCR amplification, and the forward primer is optimized RACE 5' / 3'Kit UPM, containing part of the Illumina adapter primer, the reverse primer contains the sequence of the antibody constant region and part of the Illumina adapter primer. VH, VK and VL are amplified separately, and the rea...
Embodiment 2
[0098] Example 2 Using an optimized antibody gene screening method to discover monoclonal antibodies against dengue fever
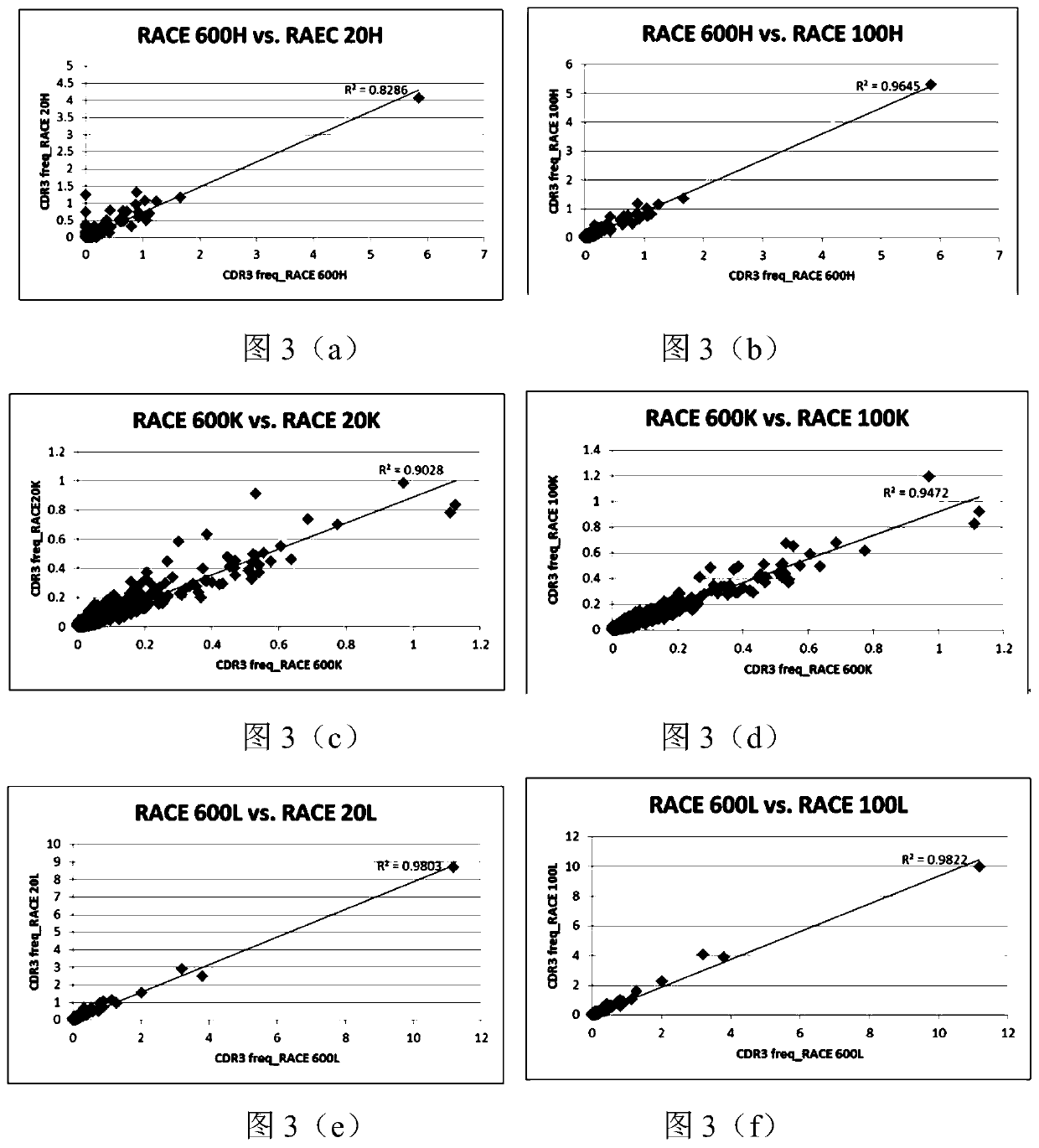
[0099] Using the RACE antibody library construction method described in Example 1, total RNA was isolated from PBMCs in the acute phase and recovery phase of dengue patients, and heavy chain, Kappa chain, and Lambda chain RACE libraries were respectively constructed. The library was sequenced using the Illumina Miseq 2×300 system, the heavy chain library was sequenced separately, and the Kappa and Lambda chain libraries were combined and sequenced. The sequencing results were processed with the software Trimmomatic-0.30, and the specific parameters were set as follows: phred33, LEADING: 20, TRAILING: 20, SLIDINGWINDOW: 20: 20, MINLEN: 200) to remove data with poor quality. The following improvements were made to the data analysis. Comparing the data volume of the antibody library with the singleton sequence removed and retained, it was found that the dat...
Embodiment 3
[0130] Example 3 Discovering Influenza Monoclonal Antibodies Using the Optimized Antibody Gene Screening Method
[0131] Using the RACE antibody library construction method described in Example 1, total RNA was isolated from PBMCs of volunteers before and 7 days after influenza vaccine injection, and the heavy chain, Kappa chain and Lambda chain RACE libraries were respectively constructed according to the method described in Example 2 ;Using Illumina Miseq 2×250 system for high-throughput sequencing. The processing of data, the method of determining FR and CDR regions, and the method of selecting candidate CDR3 amino acid sequences are all the same as those described in Example 1.
[0132] Table 10 Frequency Analysis of Heavy Chain CDR3 Before and After Influenza Vaccine Immunization
[0133]
[0134]
[0135] Table 11 Frequency analysis of Kappa chain CDR3 before and after influenza vaccine immunization
[0136]
[0137] Table 12 Frequency Analysis of Lambda Chain...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com