Method for establishing technical indicators of abundance of free and parasitic symbiotic algae of coral and coral bleaching warning coefficient H
A technical indicator and technology of symbiotic algae, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of lack of representativeness, lack of classification characteristics of symbiotic algae, interference, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] Embodiment 1: the collection of coral sample and the preparation culture test of symbiotic algae
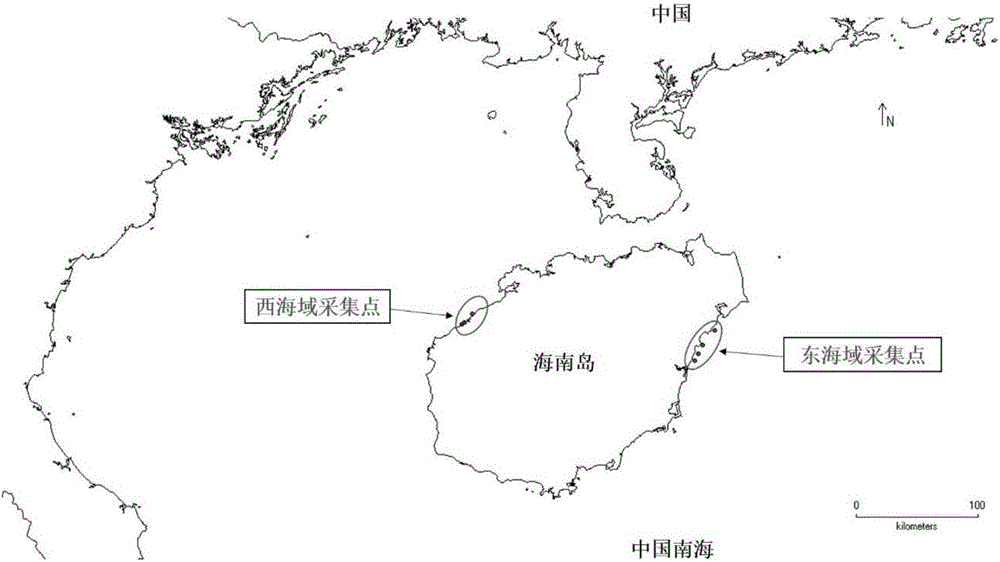
[0070] Collection of coral samples in the wild: Collect healthy helmet coral samples in the east and west waters of Hainan Island (for the GPS location of the samples, see figure 1 ), some samples were crushed or cut into small pieces, and stored in liquid nitrogen.
[0071] Collection of aquarium samples: Helmet coral samples that have been cultivated in the aquarium environment for about half a year, collected healthy and bleached coral samples, cut into small pieces, and stored in liquid nitrogen at low temperature.
[0072] In addition, 100ml of seawater samples from the aquarium were collected, and 6 repetitions were set up as the test material for detecting free symbiotic algae.
[0073] Pure culture of Symbiodinium: Coral Symbiodinium strains ITO 10 (Clade A), WZD 17 (Clade C), SGA1 (Clade D) and Symka (Clade F) were cultured under light in f / 2 medium, 27°C, photop...
Embodiment 2
[0074] The standard extraction of embodiment 2.DNA sample
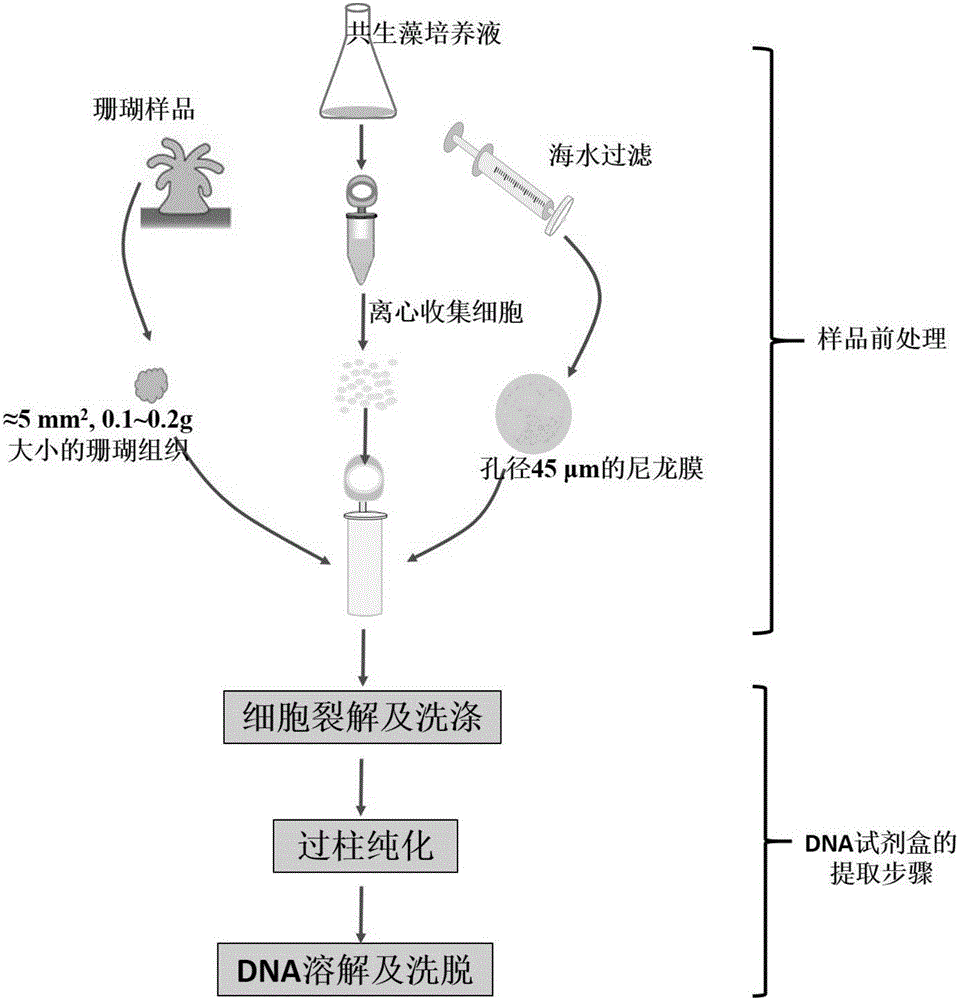
[0075] figure 2 Diagram of standard DNA extraction methods for corals, seawater samples and symbiotic algae cultures. according to figure 2 The standard extraction of DNA samples can be carried out according to the steps shown.
[0076] The marine biological DNA extraction kit (Beijing Tiangen Biotechnology Co., Ltd.) was used as the implementation of the DNA extraction method. The extraction steps are as follows: take the coral sample collected in Example 1, cut into ≈5mm 2 , 0.1-0.2g of coral tissue, take several pieces and put them into 2.0ml EP tubes, then add 500μl of lysis solution GA to obtain coral samples.
[0077] The algal cells collected by centrifugation were transferred into a 2.0ml EP tube, and 500 μl of lysate GA was added to obtain a Symbiodinium sample.
[0078] Take 500ml of seawater sample and filter it with a nylon membrane with a diameter of 0.45μm. After filtering, remove the filter membr...
Embodiment 3
[0080] Embodiment 3: the standard curve of quantitative PCR
[0081] Table 1. The combination of primers and probes for coral hosts and symbiotic algae involved in the experiment
[0082]
[0083]
[0084] Note: where Y and W are bases and combined bases, Y stands for C or T, W stands for A or T.
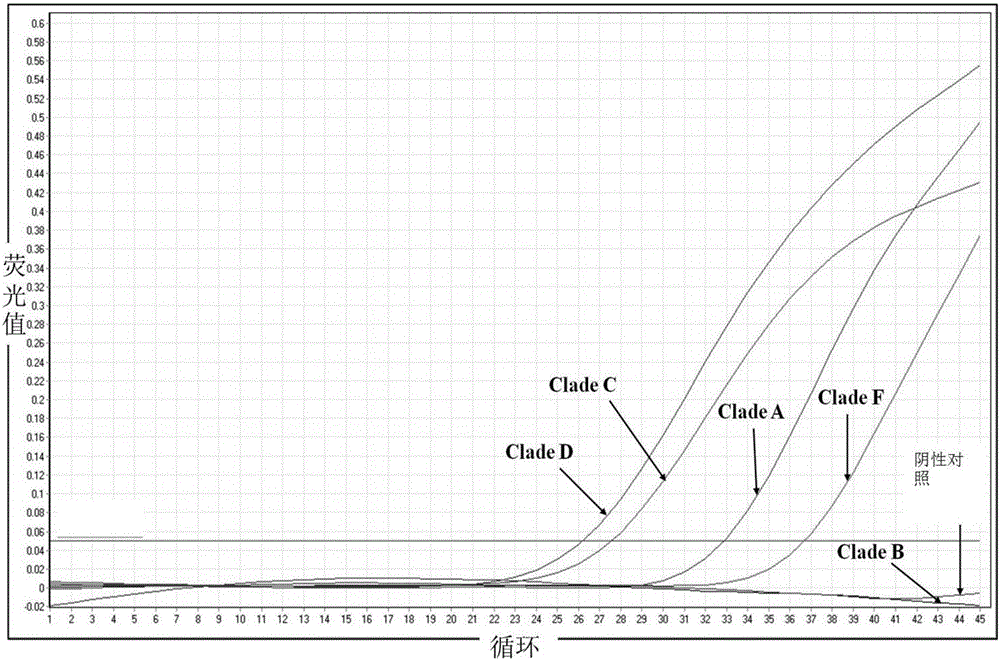
[0085] The 5' and 3' ends of the Taqman probes were labeled with a FAM reporter fluorescent dye and a TAMRA quencher, respectively. The total volume of the amplification reaction is 20 μL, including upstream and downstream primers (Forward, Reverse) 200 nM, probe (probe) 100 nM, 10 μl qPCR Probe Master Mix (Vazyme, Nanjing), template 1 μl DNA of ITO 10 (Clade A), WZD 17 (Clade C), SGA1 (Clade D) and Symka (Clade F) serially diluted by 10 times, (the above algae strains are all preserved In the China Marine Microbial Culture Collection Management Center (MCCC), MCCC is a national public welfare microbial resource sharing platform), and the rest was filled with double distille...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com