CircRNA markers for early diagnosis of lung adenocarcinoma and application thereof
An early diagnosis and marker technology, applied in the direction of recombinant DNA technology, DNA/RNA fragments, microbial detection/testing, etc., can solve the problems that circRNA has not been reported in lung cancer, and achieve broad clinical application prospects, strong specificity, and sensitivity sex high effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] 1 Clinical sample collection:
[0022] Sixty pairs of paraffin tissue specimens of early (stage IA) lung adenocarcinoma and their paired normal lung tissues were collected from Zhongda Hospital Affiliated to Southeast University during 2013-2014. All patients signed an informed consent to participate in the scientific research.
[0023] 2 circRNA quantitative PCR experiment:
[0024] 2.1 RNA extraction and quantitative PCR
[0025] Total RNA was extracted from tissues using Trizol, and RNA was quantified using a microvolume spectrophotometer.
[0026] 2.2 Primers
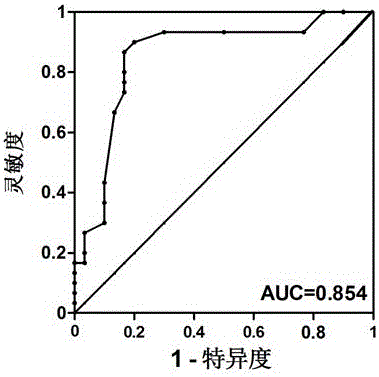
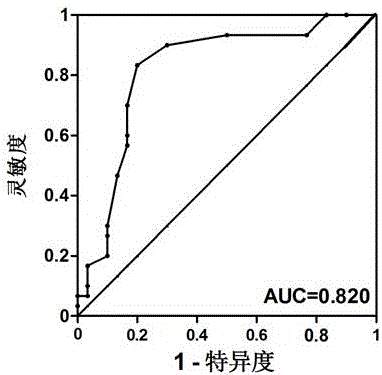
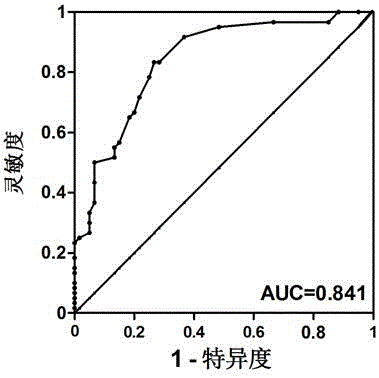
[0027] A set of circRNAs provided by the present invention includes 8 circRNAs including circRNA_100323, circRNA_69774, circRNA_403738, circRNA_404449, circRNA_104855, circRNA_100983, circRNA_1011969, and circRNA_006718. They were all selected from the results of circRNA chip detection in patients with early lung adenocarcinoma, with GAPDH as the internal reference gene (SEQ ID No: 27), and the primer seq...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com