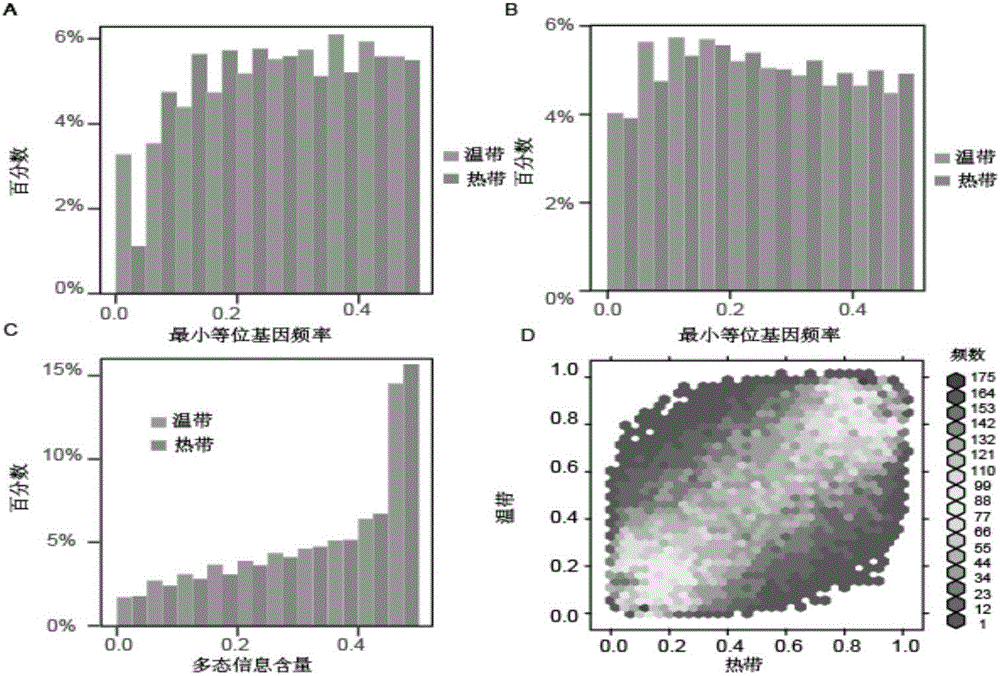
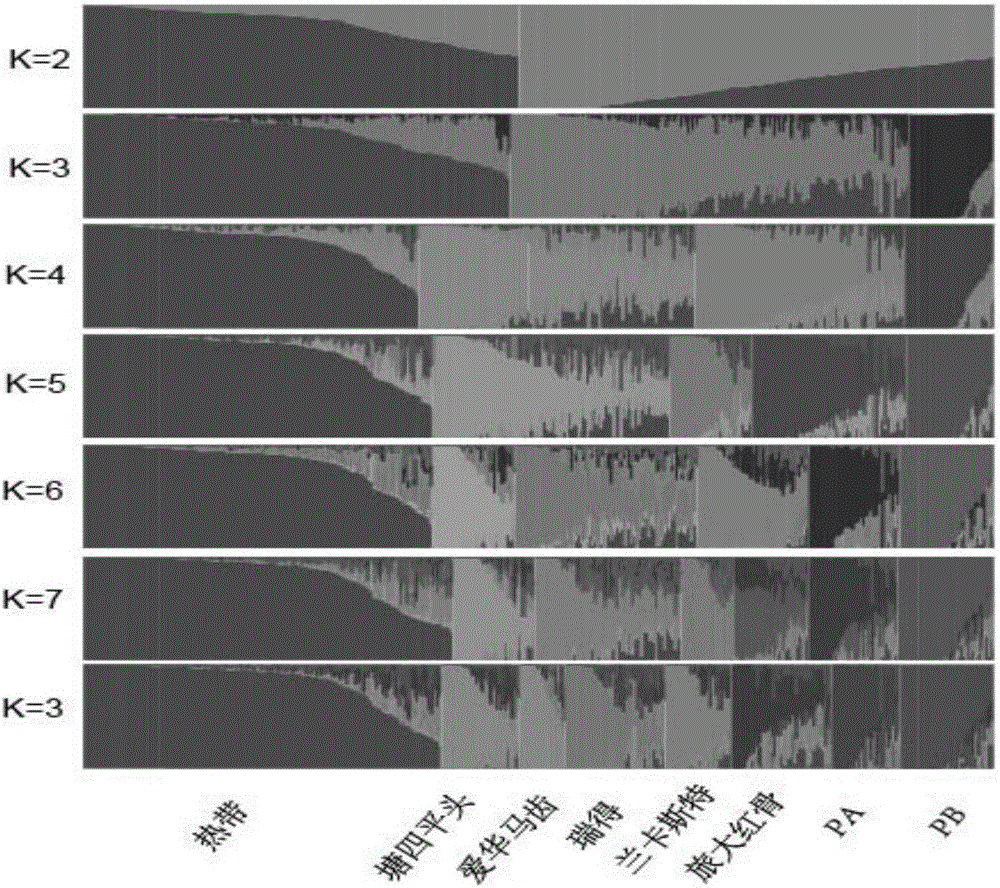
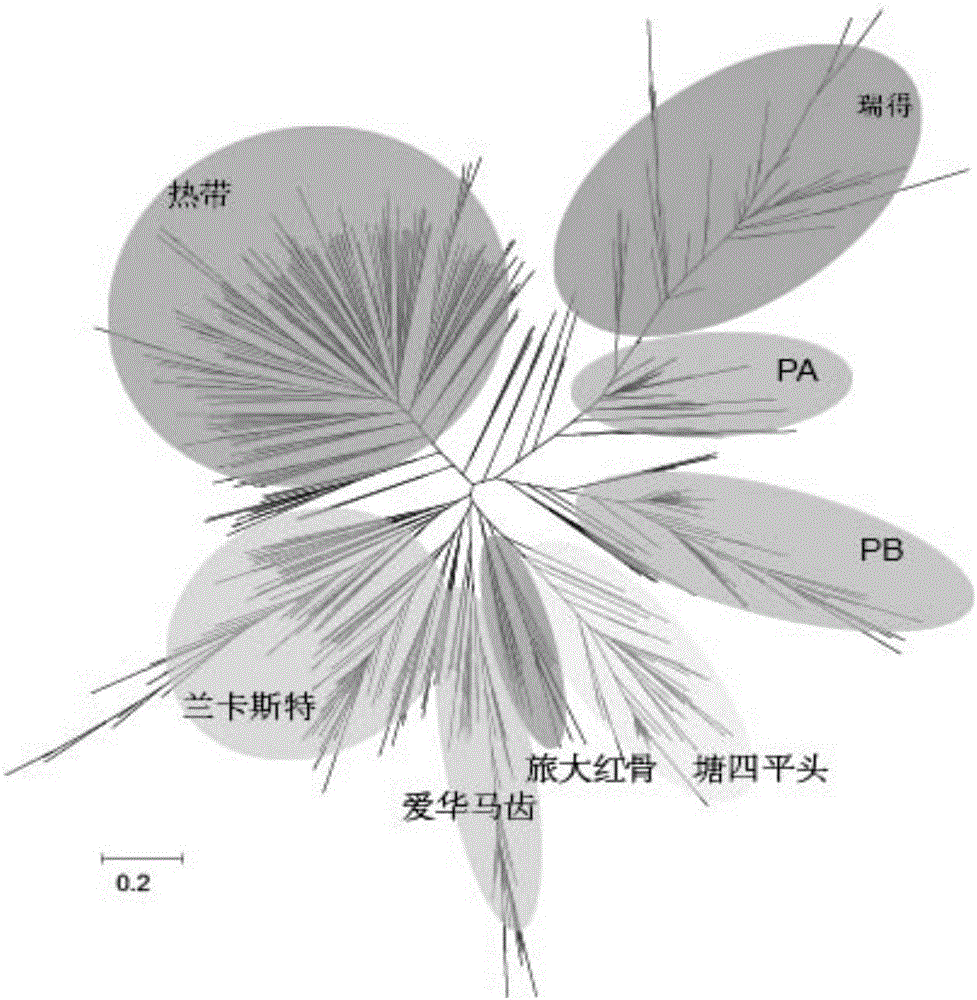
DNA chip and application of DNA chip in corn variety identification and breeding
A technology of DNA chips and DNA molecules, which is applied in the directions of DNA/RNA fragments, DNA preparation, recombinant DNA technology, etc., can solve the problems of low genome coverage, uneven distribution of chip sites, and lack of information on corn genome diversity. Effect of low locus deletion and heterozygosity, high genome coverage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1、55
[0034] Embodiment 1, the discovery process, application method and application effect of 55k chips
[0035] 1. The generation process of all SNP sites in the 55k chip
[0036] (1) Screening Europe 600K All sites in the Maize Genotyping Array were obtained to obtain 30133 SNP sites evenly distributed throughout the genome.
[0037] (2) Obtained by calculation The 10,768 loci with a deletion rate of less than 5% in MaizeSNP50BeadChip included 4,049 SNP loci with high polymorphisms in the temperate and tropical maize populations.
[0038] (3) Select 9395 SNP sites in the transcriptome sequencing data of 368 maize inbred lines.
[0039] (4) Add 451 loci related to important agronomic traits of maize.
[0040] (5) 4067 loci from the non-B73 reference genome.
[0041] (6) 734 specific loci with independent intellectual property rights for dividing heterosis groups in maize.
[0042] (7) 132 loci related to transgenic events.
[0043] After calculation, the false positive ...
Embodiment 2、1000
[0066] Embodiment 2, the development of 1000 SNP sites
[0067] When conducting maize fingerprint identification and germplasm resource evaluation, 1-3k SNP sites are most suitable considering the cost and the amount of information that the sites can provide. Based on the sequencing results of 593 maize inbred lines, the inventors of the present invention further selected a full 1000 SNPs for simplified fingerprint identification and germplasm resource evaluation. In order to select markers with uniform distribution and good polymorphism in the genome as much as possible, firstly, according to the sequencing results of 55229 SNP sites, they were divided into 1000 types. Then, based on the sequencing results of the maize inbred line B73, the entire maize genome was divided into 2000 genetic regions. Then, based on the "genetic algorithm", a group of sites is randomly selected (evenly selected according to 1000 sequencing types), and the optimal site combination that can contai...
Embodiment 3
[0068] Embodiment 3, the making and application of the chip that is used to detect 1000 SNP sites
[0069] 1. Chip preparation
[0070] The information of 1000 SNP sites is shown in Table 5.
[0071] table 5
[0072]
[0073]
[0074]
[0075]
[0076]
[0077]
[0078]
[0079]
[0080]
[0081]
[0082]
[0083]
[0084]
[0085]
[0086]
[0087]
[0088]
[0089]
[0090]
[0091]
[0092]
[0093]
[0094] Note: The probes are all single-stranded DNA molecules.
[0095] The probes described in Sequence 1 to Sequence 1000 are respectively solidified on the same substrate in the form of an array to obtain a specific DNA chip (AXIOM gene chip).
[0096] 2. Chip application
[0097] The samples to be tested are: 593 corn inbred lines (the eight corn inbred lines are: corn inbred line H201, corn inbred line Ji 046, corn inbred line Ji495, corn inbred line K14, corn inbred line line B73, corn inbred line CML2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com