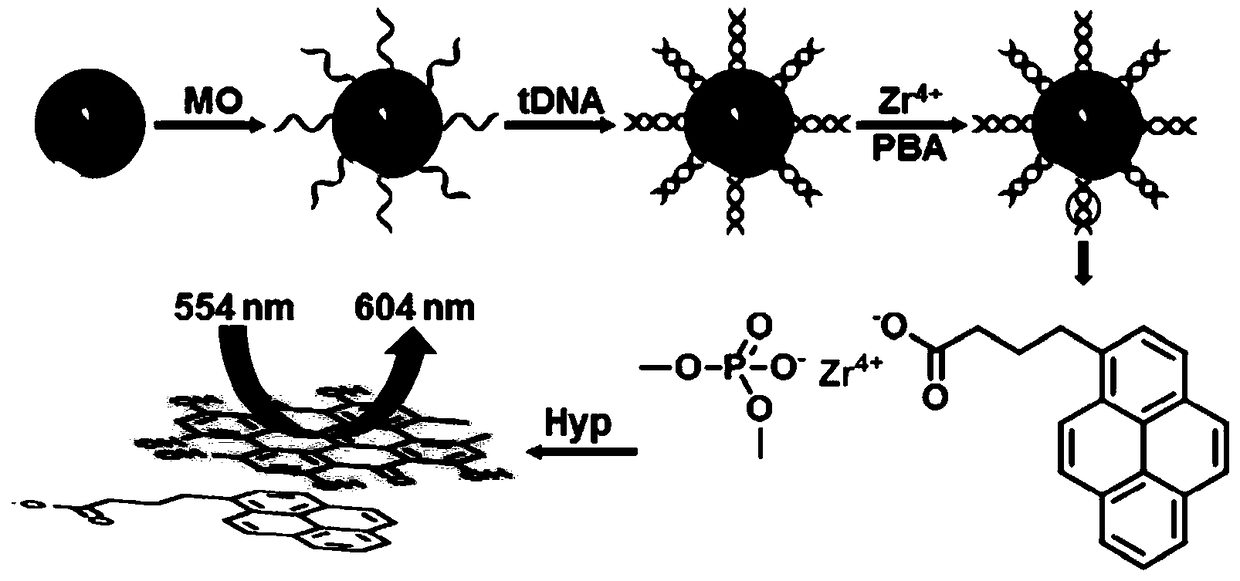
DNA fluorescence analysis method based on the interaction between hypericin and 1-pyrene butyric acid
A fluorescence analysis and magnetic separation technology, applied in the field of DNA fluorescence analysis, can solve the problems of difficult single nucleotide polymorphism detection, high background noise of electrochemical method, low hybridization efficiency, etc., achieve good linear response, simple method, The effect of short analysis times
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
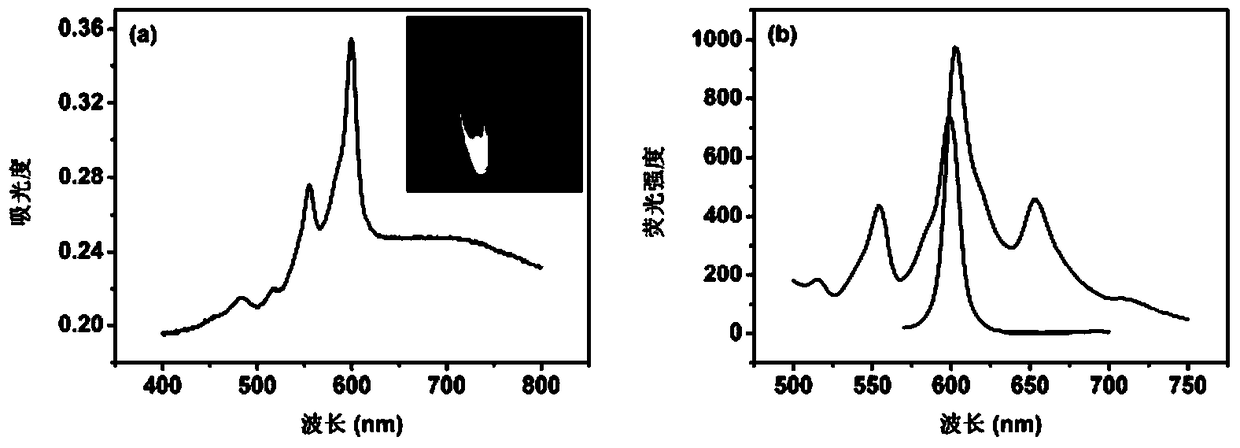
[0029] Example 1: Ultraviolet and fluorescence spectrum characterization of fluorescent probes introduced on the surface of SA-MB through the coordination of zirconium ions and the π-π stacking between Hyp and PBA.
[0030] Using the DNA fluorescence analysis based on the π-π stacking effect between Hyp and PBA described in the present invention, suspend 0.5 mg / mL SA-MB in PBS buffer, add 1 μM MO modified with biotin at the 3' end, The sequence is 5'-AAC CAT ACA ACCTAC TAC CTC A-Biotin-3'. After reacting for 60 minutes, perform magnetic separation and wash with PBS buffer several times, then add 100nM tDNA (dissolved in TE buffer), the sequence is 5'- TGA GGT AGT AGG TTG TAT GGT T-3', after hybridization for 60min, perform magnetic separation and wash with Tris-HCl buffer several times, then add 4mM ZrOCl 2 ·8H 2 O solution (dissolved in 60% ethanol), after coordination reaction for 30min, carry out magnetic separation and wash with 60% ethanol several times, then add 2mM PBA...
Embodiment 2
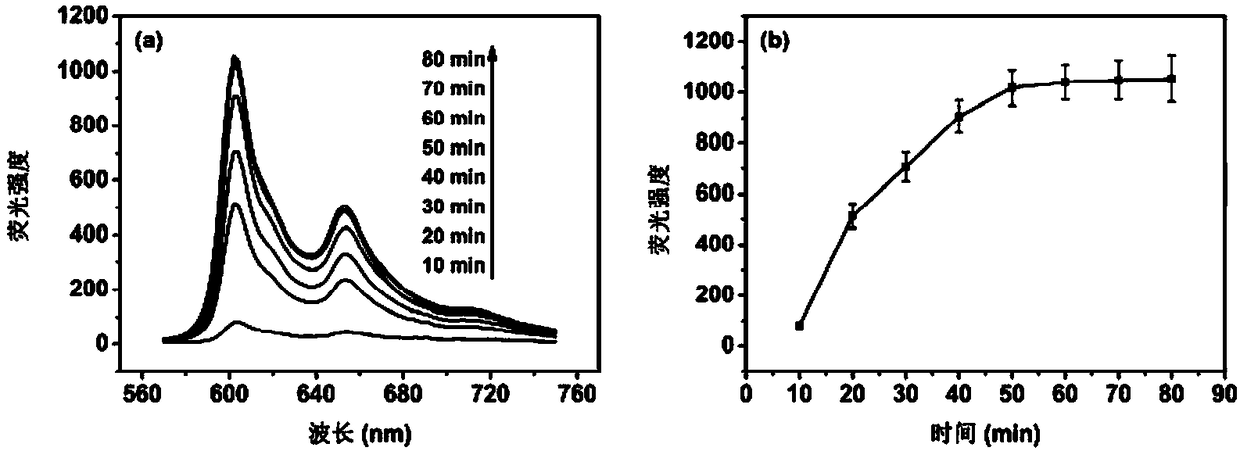
[0031] Example 2: Time optimization of π-π stacking interaction between Hyp and PBA.
[0032] Using the DNA fluorescence analysis based on the π-π stacking interaction between Hyp and PBA according to the present invention, the operation steps are the same as the above-mentioned Example 1. Among them, 100nM tDNA fragment with the sequence of 5'-TGA GGT AGT AGG TTG TAT GGT T-3' was selected as the target analyte, and the π-π stacking time was changed to 10, 20, 30, 40, 50, 60, 70 , 80min, analyze the influence of reaction time on the DNA fluorescence analysis result of the present invention, its analysis result is as attached image 3As shown, where a is the change of the fluorescence spectrum, b is the change of the peak value, it can be seen from the figure that when the π-π stacking time is 10, 20, 30, 40, 50 min, the fluorescence intensity increases significantly with the increase of time , and after 50 min of reaction, the fluorescence intensity remained stable with the i...
Embodiment 3
[0033] Example 3: Effect of water content on π-π stacking.
[0034] Using the DNA fluorescence analysis based on the π-π stacking interaction between Hyp and PBA according to the present invention, the operation steps are the same as the above-mentioned Example 1. Among them, 100nM of the tDNA fragment with the sequence of 5'-TGA GGT AGT AGG TTG TAT GGT T-3' was selected as the target analyte, and the water content in the π-π stacking reaction was changed to 0%, 10%, 20%, 30% %, 40%, 50%, 60%, 70%, 80%, 90%, 100%, to analyze the effect of water content on π-π stacking, the analysis results are as attached Figure 4 As shown, where a is the comparison of fluorescence spectra, and b is the comparison of peak values. It can be seen from the figure that with the increase of water content, the fluorescence intensity decreases significantly. After the water content exceeds 80%, the fluorescence intensity almost drops to zero, so in In the actual detection process, it is necessary t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com