Method for detecting transcription factors on basis of cooperative masking effect and binding protection effect mediation cascade signal amplification strategies
A technology of transcription factors and reactions, applied in biochemical equipment and methods, microbiological determination/inspection, DNA/RNA fragments, etc., can solve problems such as false positive signals, and achieve the effect of eliminating false positive signals
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
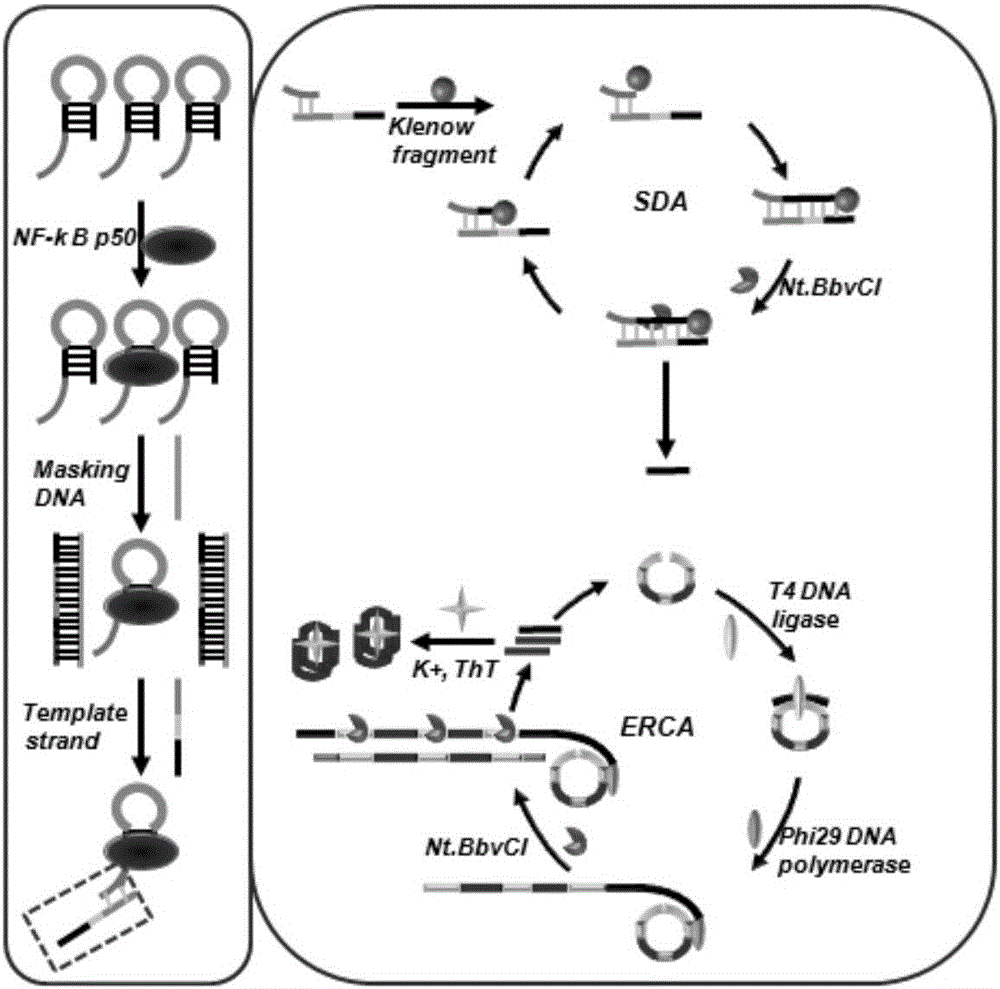
[0062] Example 1: Detection of transcription factors (NF-κB p50) based on a cascade signal amplification strategy mediated by cooperative masking effects and binding protective effects
[0063] The specific method is as follows:
[0064] (1) Binding of target and hairpin recognition probe
[0065] 1 μL Cutsmart, 40 nM hairpin recognition probe, different concentrations of NF-κBp50 or actual samples were included in a 10 μL reaction system, the volume of the reaction system was 10 μL, and placed in a 37°C incubator after slight shaking for 3 hours.
[0066] (2) masking reaction
[0067] The 11 μL reaction system contained 10 μL of the binding product, 1 μM DNA masking strand, placed in a 37 ° C incubator after slight shaking, and reacted for 0.5 h.
[0068] (3) Strand Displacement Amplification (SDA)
[0069] In a 20 μL reaction system, 11 μL masked reaction product, 2 μL Cutsmart, 1.5 mM dNTPs, 1UKlenow fragment, 2U Nt.BbvCI, placed in a 37°C incubator after slight shaking,...
Embodiment 2
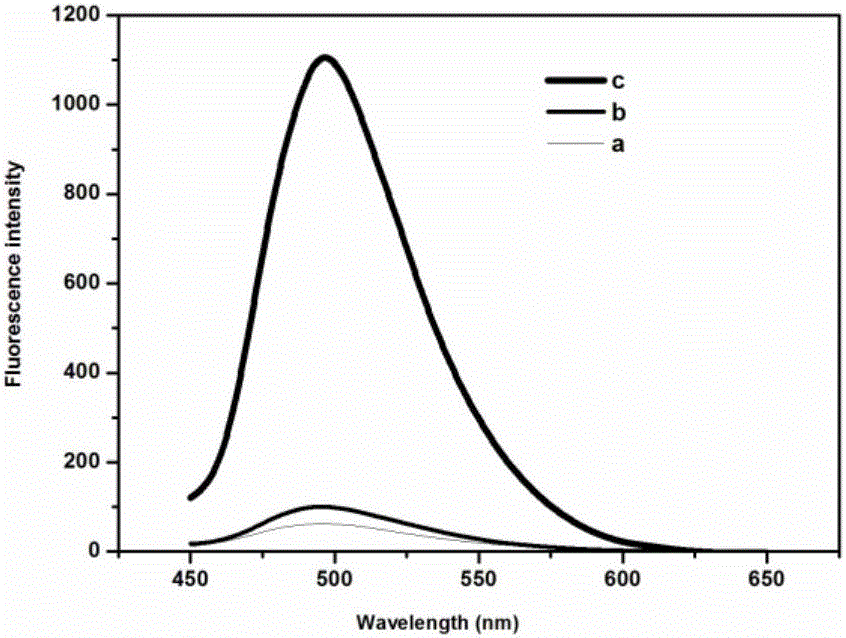
[0075] Embodiment 2: Feasibility study of detection method of the present invention
[0076] In order to verify the feasibility of the detection method of the present invention, the present invention examines the feasibility of the novel detection strategy proposed herein by measuring the fluorescence intensity under different conditions, such as figure 2 shown. It can be seen that the dyes ThT (curve a) and negative (curve b) show relatively weak fluorescence, indicating that in the absence of the target NF-κB p50, the masking strand hybridizes with the recognition probe to form a stable double-stranded structure, with purple ends Enclosed in the double strand, it cannot hybridize with the template strand, triggering the subsequent cascade of signal amplification. When the target exists, the fluorescence intensity is significantly enhanced (curve c), indicating that the target binds to the recognition probe to form a complex, and the steric hindrance effect prevents the mas...
Embodiment 3
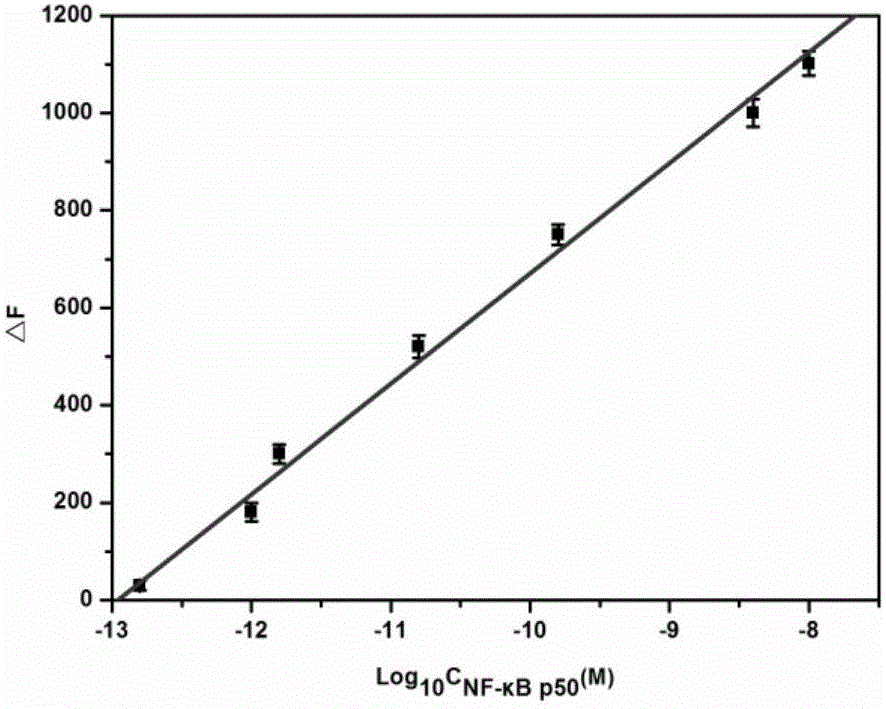
[0077] Embodiment 3: the sensitivity investigation of detection method of the present invention
[0078] The linear range and sensitivity of the method can be determined by measuring the fluorescence intensity corresponding to different concentrations of the target. Such as image 3 shown. It can be seen that the target concentration is 1.5×10 -13 M-1.0×10 -8 In the M range, ΔF has a linear relationship with the logarithm of the target concentration (R 2=0.993), the estimated LOD of this detection method is 1.0×10 -13 M, better than most reported sensitivities in the literature.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com