Kit and method for detecting activity of DNA methyltransferase
A kit and methylation technology, applied in the field of biological analysis, can solve the problems of low sensitivity, simple instrument, non-specific exponential amplification, etc., and achieve the effect of high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
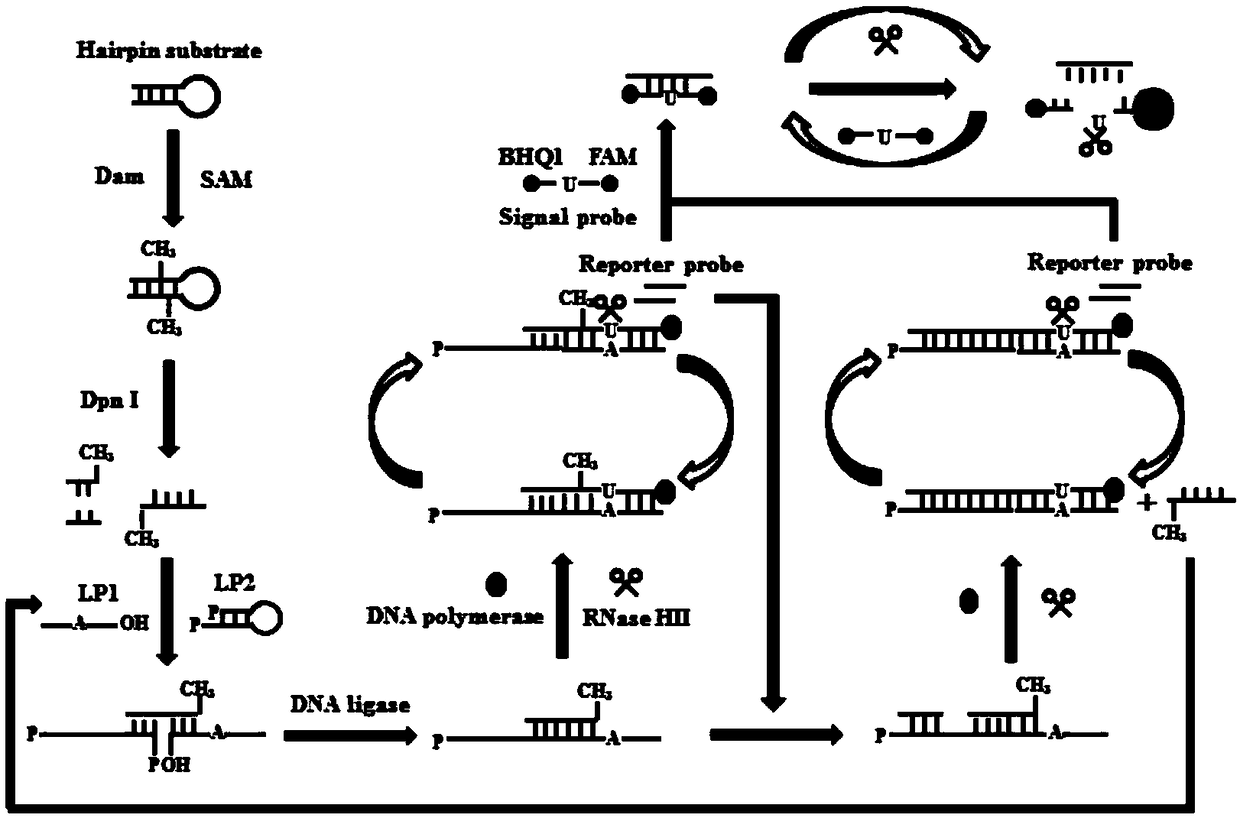
[0083] Circular junction-dependent signal amplification mediated by single ribonucleotide repair: detection of DNA methyltransferase (MTase) involves two sequential reaction steps. First, dilute all oligonucleotides with 1× Tris-EDTA buffer (10 mmol / L Tris, 1 mmol / L ethylenediaminetetraacetic acid (EDTA), pH 8.0) Acid sequence to prepare stock solutions. Then, the hairpin substrate and ligation probe 2 (LP2) were dissolved in buffer (1.5 mmol per liter of magnesium chloride, 10 mmol per liter of tris(hydroxymethyl)aminomethane-hydrochloric acid (Tris-HCl), pH 8.0) to 10 micromolar per liter and incubated at 95°C for 5 minutes, then slowly cooled to room temperature to form a hairpin structure. Subsequently, 1 µl of hairpin substrate (10 µmol / L) was added to 20 µl containing Dam MTase, 160 µmol / L of SAM, 6 units of Dpn I, 2 µl of 10×Dam MTase Reaction buffer and 2 μl of 10×CutSmart buffer were incubated at 37°C for 2 hours, then inactivated at 80°C for 20 minutes. Then 5 mic...
Embodiment 2
[0088] (1) Experimental verification of the principle:
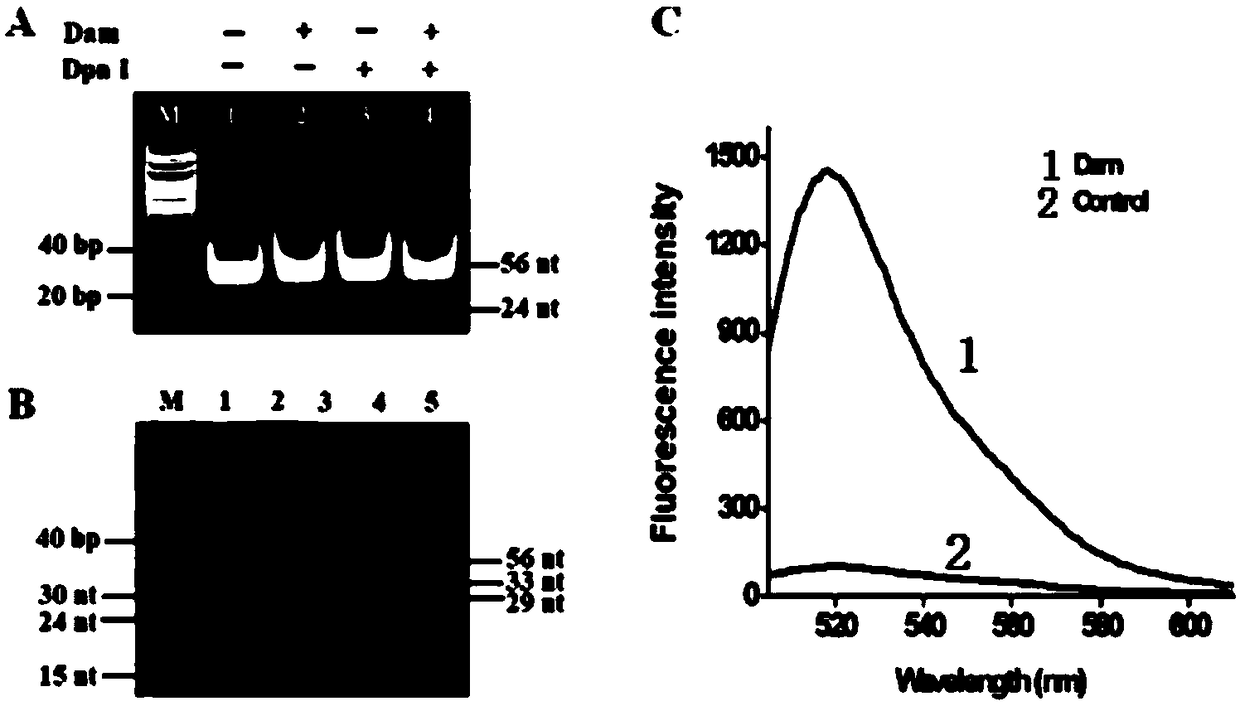
[0089] In order to verify whether Dam MTase can cleave hairpin substrates in the presence of Dpn I, we used non-denaturing polyacrylamide gel electrophoresis and used SYBR Gold as a fluorescent indicator to analyze the reaction products ( figure 2 A). When both Dam MTase and Dpn I do not exist ( figure 2 A, lane 1), only Dam MTase ( figure 2 A, lane 2) and only Dpn I ( figure 2 When A lane 3) was present, only one 56-nt band was observed, which was the same size (56 nt) as the synthetic hairpin substrate, indicating that no methylation and cleavage reactions occurred. But when in the presence of Dam MTase and Dpn I, a new 24-nt band was observed ( figure 2 A, lane 4), indicating that Dam MTase can trigger the methylation of the hairpin substrate, and then the methylated hairpin substrate can be cleaved by Dpn I at the methylated adenine site.
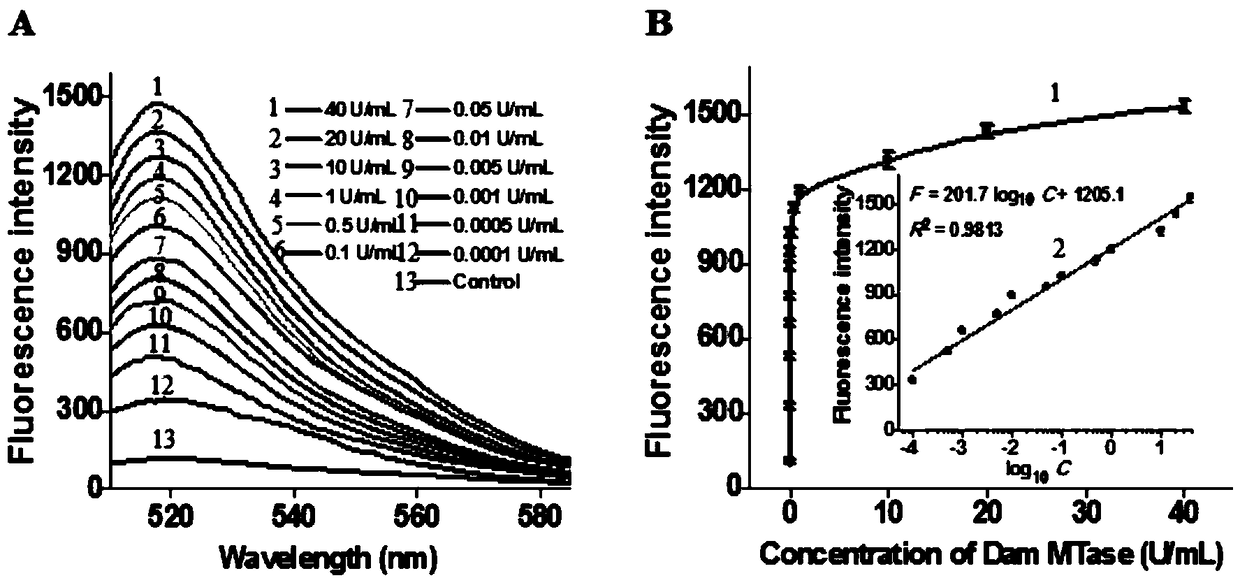
[0090] In order to verify the cyclic junction-dependent SDA reactio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com