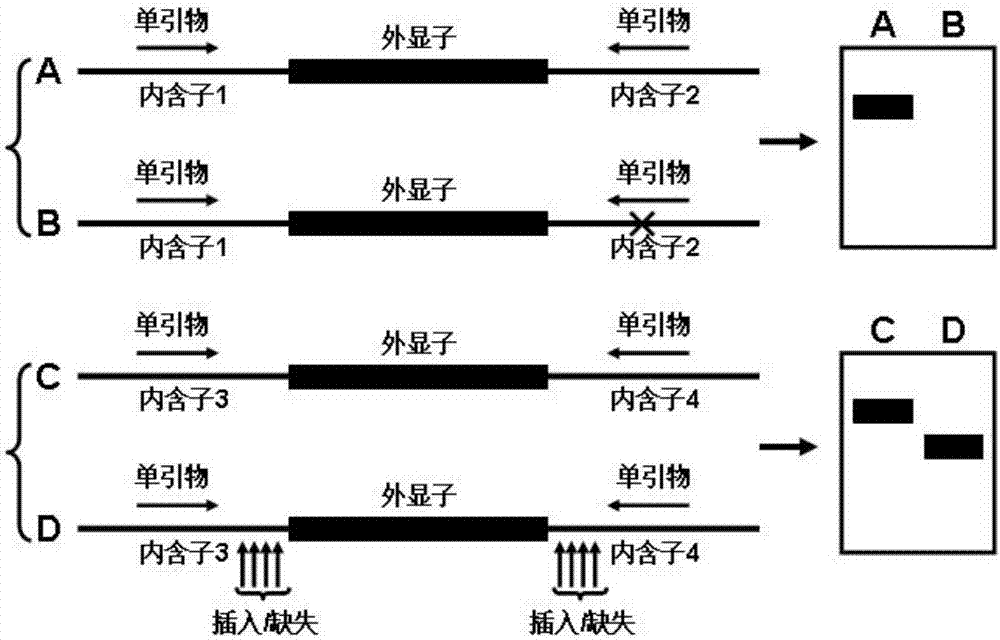
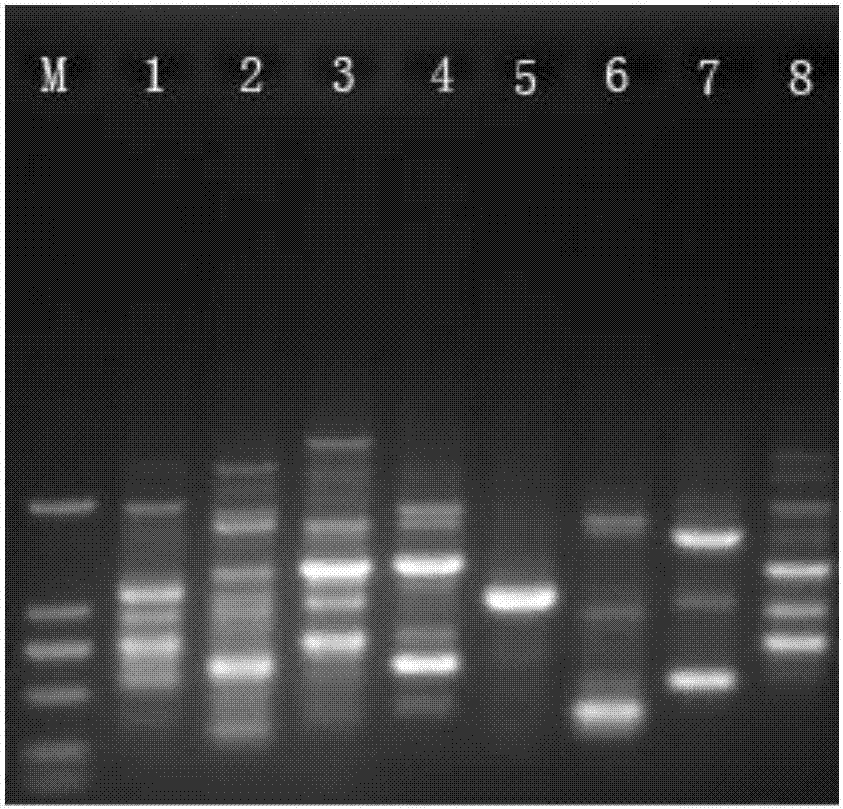
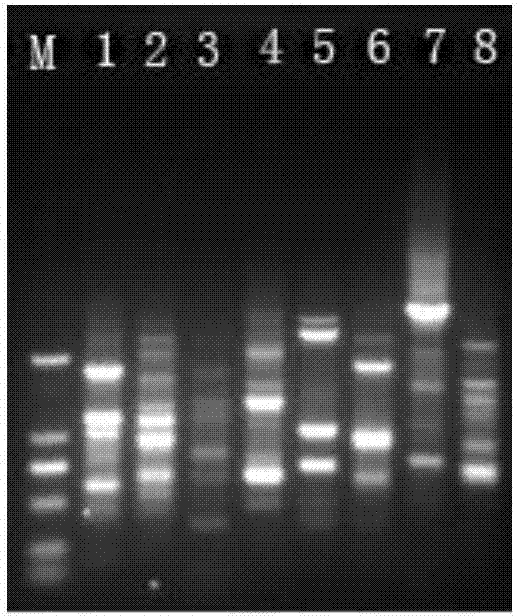
Single primer capable of multi-species molecular marking and marking method thereof
A technology of molecular markers and marker methods, which is applied in biochemical equipment and methods, measurement/inspection of microorganisms, DNA/RNA fragments, etc., and can solve the problems of undisclosed multi-species molecular marker analysis, poor repeatability of primer amplification, and primer annealing temperature. Low-level problems, to achieve the effect of convenient molecular marker-assisted selection breeding, rich amplification bands, and rich polymorphisms
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0084] Example 1: In this example, peanuts are used as samples to verify the effectiveness of single primer amplification and screening experiments
[0085] 1. Sample material: Guihua 911 (temporary name), a new peanut strain.
[0086]2. Peanut genomic DNA extraction: Take 0.3-0.5g of fresh peanut leaf samples, cut them into pieces with scissors, put them into a mortar and add 1g of PVP to the mortar, pour enough liquid nitrogen into the mortar several times, and put The leaf samples were fully ground into powder, and all the powder samples were transferred to a 2ml centrifuge tube that had been preheated at 65°C with 1.0ml of CTAB extract in advance, then added 20μl of β-mercaptoethanol, and incubated in a water bath at 65°C for 60 minutes. Mix upside down so that the DNA can be fully separated; then centrifuge at 12000rpm in a refrigerated centrifuge for 10 minutes to remove the bottom residue, draw the supernatant to another 2ml centrifuge tube, and add 0.5μl of RNaseA to t...
Embodiment 2
[0092] Example 2: In this example, sugarcane is used as a sample to verify the effectiveness of single primer amplification and screening experiments
[0093] 1. Sample material: sugarcane variety New Taiwan Sugar 22.
[0094] 2. Sugarcane genomic DNA extraction: Take 0.3-0.5g of fresh sugarcane core leaves, cut them into pieces with scissors, put them into a mortar and add 1g of PVP to the mortar, pour enough liquid nitrogen into the mortar several times, and put the leaves The sample is fully ground into powder, and all powder samples are transferred to a 2ml centrifuge tube that has been preheated at 65°C with 1.0ml of CTAB extract in advance, then added 20μl of β-mercaptoethanol, and warmed in a water bath at 65°C for 60 minutes. Invert and mix well so that the DNA can be fully separated; then centrifuge in a refrigerated centrifuge at a speed of 12000rpm for 10 minutes to remove the bottom residue, draw the supernatant to another 2ml centrifuge tube, and add 0.5 μl of RNa...
Embodiment 3
[0097] Example 3: In this example, rice is used as a sample for molecular marker analysis
[0098] 1. Sample material: 16 rice varieties, namely: Kasalah, Baipi Nuogu, Wandaonuo, Huagu, Taiwanbai, Xigu, Huangbaizhan, Guanghui 998, Nipponbare, Huanghongnuo, Xiangnuo, Shanjaponica , Malay, Huapi, Late Upland Rice and Rongbaohong.
[0099] 2. Genomic DNA extraction: Take 0.3-0.5g of fresh rice tender leaves, cut them into pieces with scissors, put them into a mortar and add 1g of PVP to the mortar, pour enough liquid nitrogen into the mortar several times, and the leaf samples Fully grind into powder, transfer all powder samples to a 2ml centrifuge tube that has been preheated at 65°C with 1.0ml of CTAB extract in advance, then add 20μl of β-mercaptoethanol, and warm it in a water bath at 65°C for 60 minutes, during which time it is turned upside down Mix well so that the DNA can be fully separated; then centrifuge at 12000rpm in a refrigerated centrifuge for 10 minutes to remov...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com