Method for rapidly identifying H. villosa chromosomes in wheat background
A tufted wheat and chromosome technology, applied in the field of molecular cytogenetics, can solve the problems of inability to identify rye chromosomes, complicated processes, and inability to be clear at a glance, and achieves the effects of overcoming cumbersome preparation, low cost and easy operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1 Analysis of Wheat-Viligal repeats and Determination of Oligonucleotide Probe Sequences
[0049] According to the characteristics of the distribution of transposable repeat sequences in the plant genome in a scatter manner, we start with the analysis of transposable repeat sequences and search for oligonucleotide probes. Based on the fact that there are relatively few transposable repetitive sequences isolated and cloned from the genome of T. villosa, and based on the principle that each species of wheat crops has a certain genetic relationship, consider using the method of sequence transfer between species to develop suitable oligonucleotides acid probe.
[0050] 1. Determination of oligonucleotide probe nucleotide sequence and probe design
[0051] (1) Collect all the reported transposable repeat sequences isolated and cloned from the rye genome and the reported rye-specific oligonucleotide probe sequences.
[0052] From the NCBI database, download all publ...
Embodiment 2
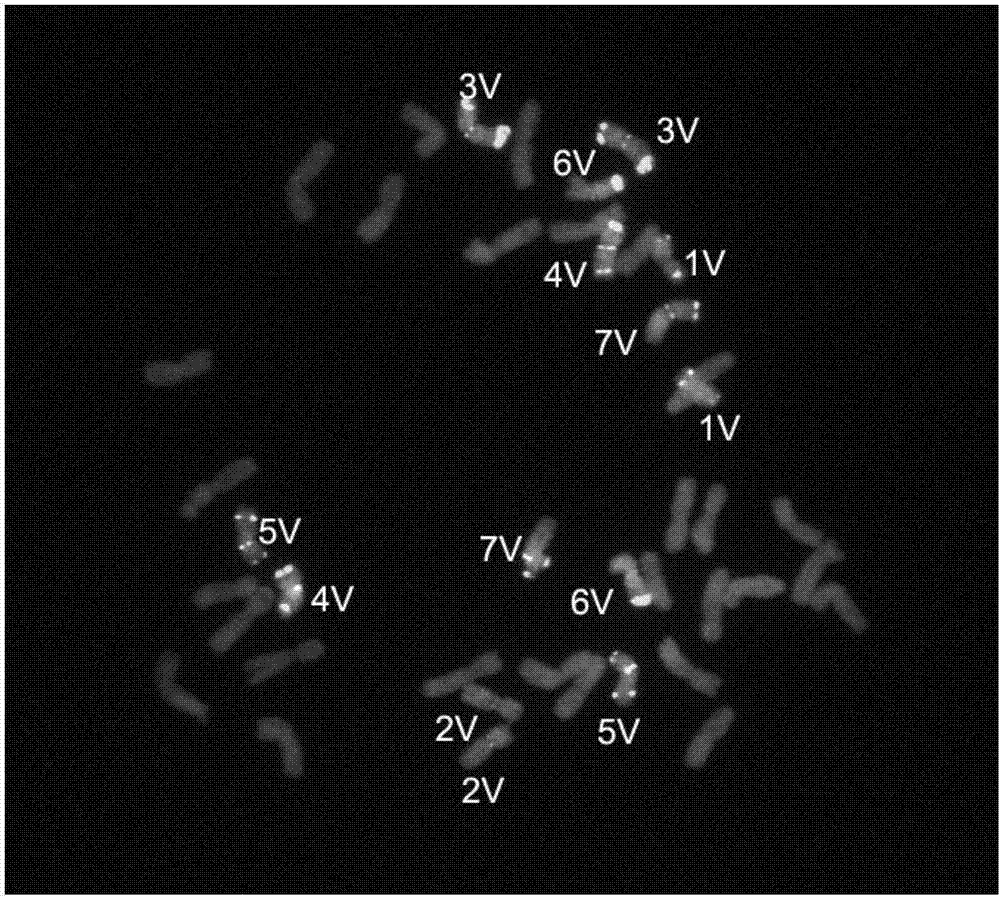
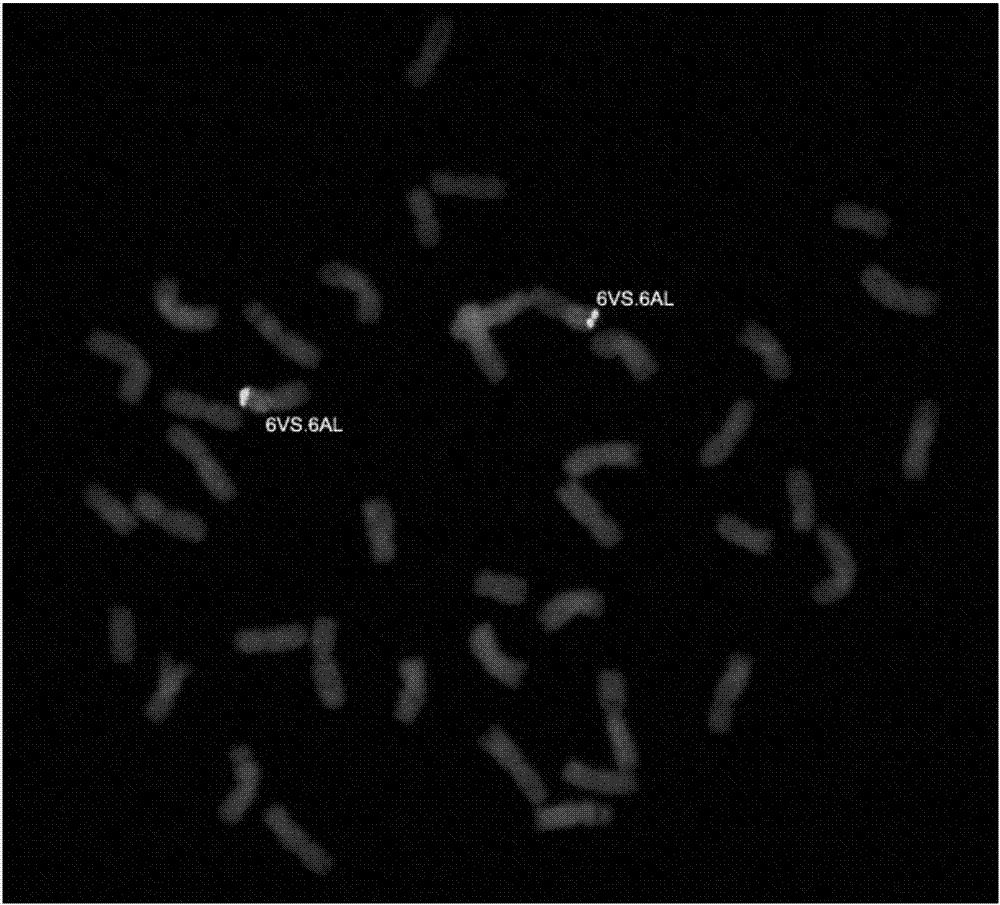
[0060] Example 2 Establishment of the Chromosomal Identification Method for T. villosa in the Wheat Background
[0061] Two kinds of oligonucleotide probes (Oligo-Ku and (GT) determined in embodiment 1 7 ) were mixed, and ND-FISH was used to identify the chromosomes of A. villosa in the wheat background. The method described in (Han et al.2006) was used for the preparation of the root tip metaphase chromosomes of wheat-T. villosa amphidiploid and wheat cultivar Mianmai 37, and the method described in (Fu et al.2015) for the ND-FISH analysis process . details as follows:
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com